A PIPELINE FOR LUNG TUMOR DETECTION AND SEGMENTATION FROM CT SCANS USING DILATED CONVOLUTIONAL NEURAL NETWORKS
DOI: 10.1109/ICASSP.2019.8683802
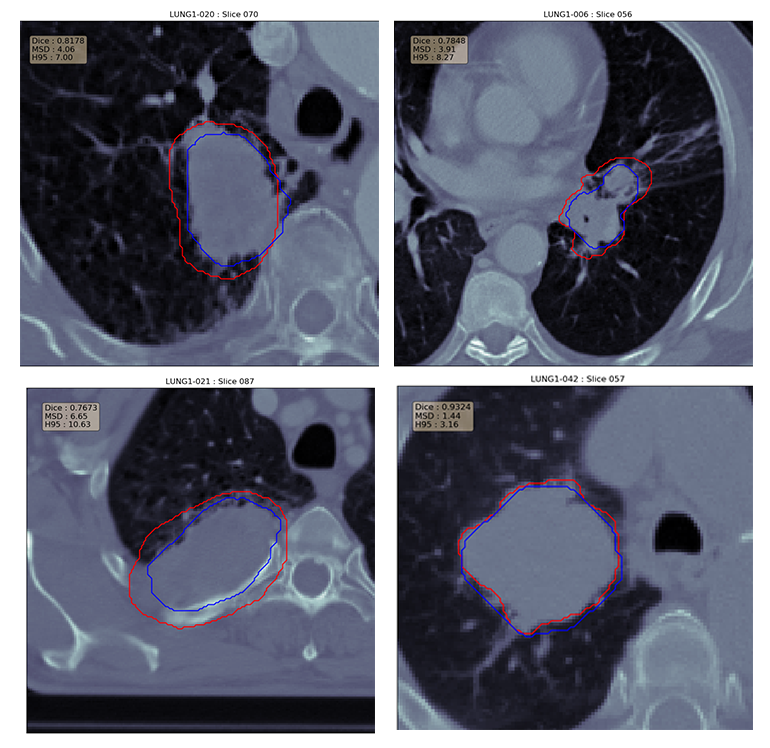
Lung cancer is the most prevalent cancer worldwide with about 230,000 new cases every year. Most cases go undiagnosed until it’s too late, especially in developing countries and remote areas. Early detection is key to beating cancer. Towards this end, the work presented here proposes an automated pipeline for lung tumor detection and segmentation from 3D lung CT scans from the NSCLC Radiomics Dataset. It also presents a new dilated hybrid-3D convolutional neural network architecture for tumor segmentation. First, a binary classifier chooses CT scan slices that may contain parts of a tumor. To segment the tumors, the selected slices are passed to the segmentation model which extracts feature maps from each 2D slice using dilated convolutions and then fuses the stacked maps through 3D convolutions - incorporating the 3D structural information present in the CT scan volume into the output. Lastly, the segmentation masks are passed through a post-processing block which cleans them up through morphological operations. The proposed segmentation model outperformed other contemporary models like LungNet and U-Net. The average and median dice coefficient on the test set for the proposed model were 65.7% and 70.39% respectively. The next best model, LungNet had dice scores of 62.67% and 66.78%.
- NSCLC-Radiomics Dataset - 3D CT Scans with tumor regions manually annotated
| Set | Patients | With Tumor | Without Tumor |
|---|---|---|---|
| Train | 260 | 4,296 | 26,951 |
| Test | 40 | 848 | 3,630 |
- X/Y Mirroring
- Rotations
- Elastic Transformation
- 7 fold increase in data
Three subsets created from total data pool:
- A: all Tumor Slices only
- B: A + 10 non-tumor slices from each patient
- C: 3D stacks of 9 consecutive slices
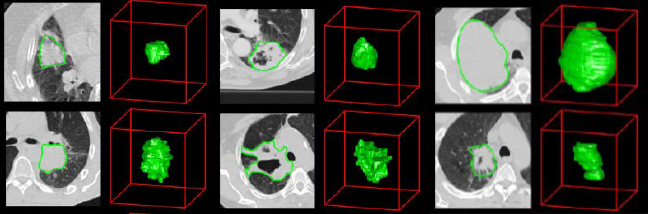 Sample Slices from the NSCLC Dataset
Sample Slices from the NSCLC Dataset
- Detecting and Cropping out Lung Region (Image Processing)
- Adaptive Histogram Equalization
- Normalization and Resizing
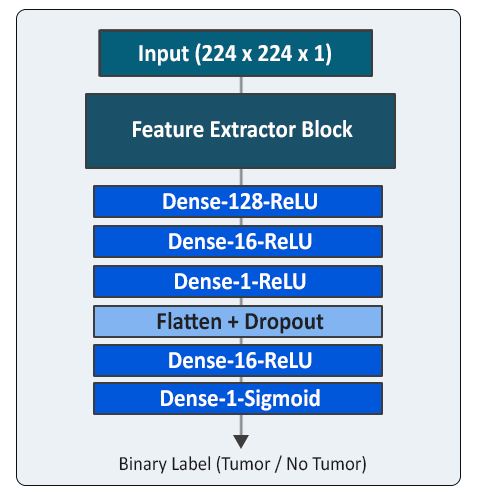
- Feature Extraction from each 2D slice using LungNet2D
- Binary Classifier labels probable slices containing tumor
- Tumors generally localized in small volume (3-30 slices = 10 ~ 100 mm3) compared to total lung volume ( ~ 900 mm3)
- Frontend binary classifier used to weed out non tumor containing slices
- Positive detection = 8 neighboring slices along the ±z directions passed to Segmentation Model
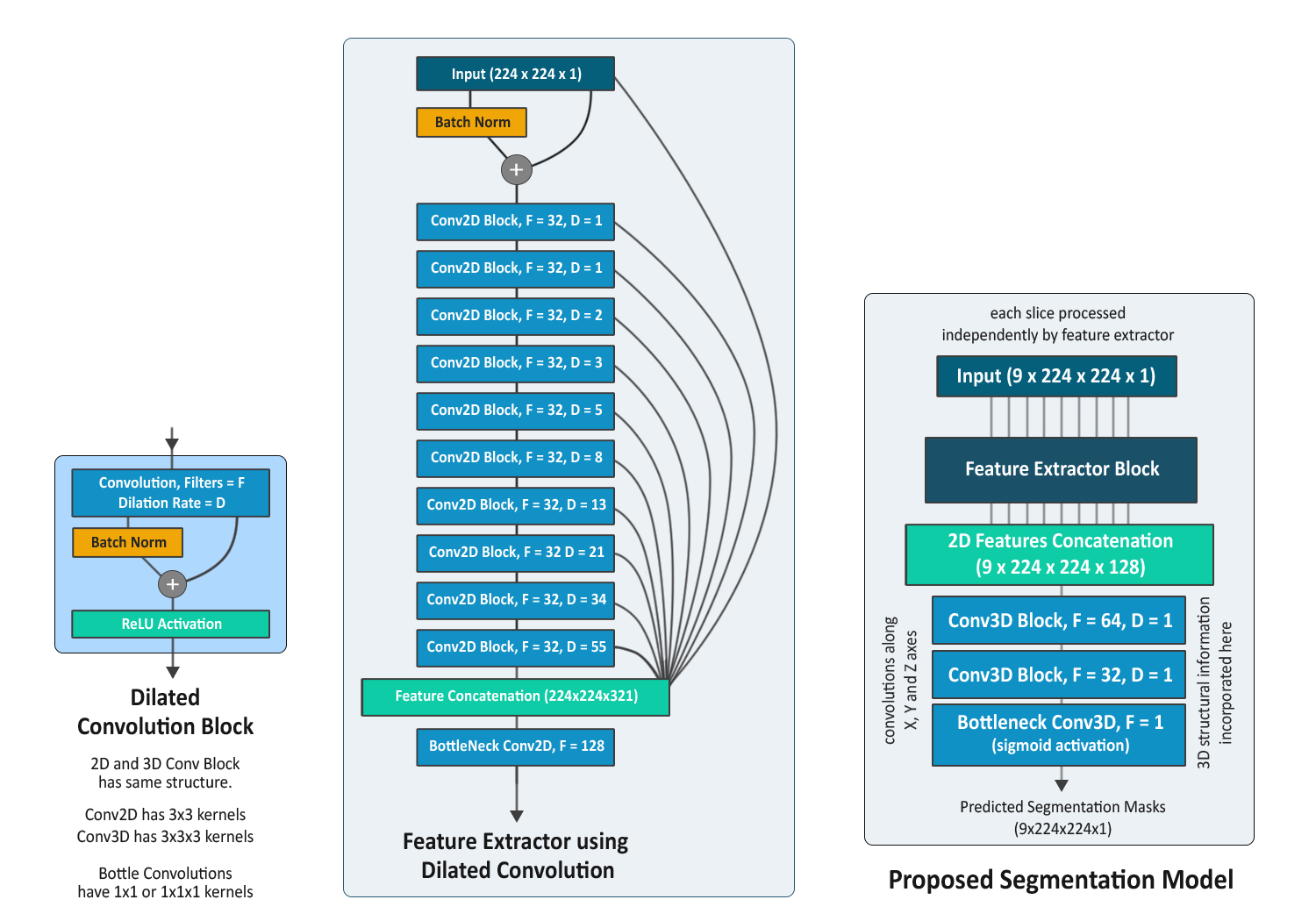
- Feature Extraction Block (FEB) = LungNet2D
- Dilation Rate increased in Fibonacci Sequence
- Initially trained with 2D slices to output 2D masks
- FEB applied to each of the 9 slices in mini stack
- Feature maps are fused through 3D Convolution
- Outputs 3D masks
- Area based Thresholding to get rid of specks (region < 5mm2 removed)
- Morphological Operations to refine segmentation mask
- LungNet2D trained first on subset A and then subset B on decimated learning rate
- To transform LungNet2D into the proposed model, LungNet3D
- Final Convolutional layers switched to 3D convolutions
- Instead of single slice input, stacks of 9 slices provided
- LungNet3D trained on subset C
- The trained LungNet2D model was used as base
- Final Convolutional Layers replaced by fully connected layers
- Trained on subset B to output binary labels (tumor/non-tumor)
| Model Arch. | Total params (x10^6) | Mean Dice (%) | Median Dice (%) |
|---|---|---|---|
| U-NET | 31 | 58.5 | 62.3 |
| LungNet 2D | 0.13 | 62.7 | 66.8 |
| Proposed | 0.40 | 65.8 | 70.4 |