-
Notifications
You must be signed in to change notification settings - Fork 103
Home
- Summary
- System requirements
- Memory requirements
- Installation
- Getting started
- Frequently Asked Questions
Foldseek runs on modern UNIX operating systems and is tested on Linux and macOS. Additionally, we are providing a preview version for Windows.
Foldseek takes advantage of multi-core systems through OpenMP and uses the SIMD capabilities of the system. Optimal performance requires a system supporting the AVX2 instruction set, however SSE4.1 and very old systems with SSE2 are also supported. It also supports the PPC64LE and ARM64 processor architectures, these require support for the AltiVec or NEON SIMD instruction sets, respectively.
To check if Foldseek supports your system execute the following commands, depending on your operating system:
[ $(uname -m) = "x86_64" ] && echo "64bit: Yes" || echo "64bit: No"
grep -q avx2 /proc/cpuinfo && echo "AVX2: Yes" || echo "AVX2: No"
grep -q sse4_1 /proc/cpuinfo && echo "SSE4.1: Yes" || echo "SSE4.1: No"
# for very old systems which support neither SSE4.1 or AVX2
grep -q sse2 /proc/cpuinfo && echo "SSE2: Yes" || echo "SSE2: No"
[ $(uname -m) = "x86_64" ] && echo "64bit: Yes" || echo "64bit: No"
sysctl machdep.cpu.leaf7_features | grep -q AVX2 && echo "AVX2: Yes" || echo "AVX2: No"
sysctl machdep.cpu.features | grep -q SSE4.1 && echo "SSE4.1: Yes" || echo "SSE4.1: No"
To ensure optimal performance of the software, it is important to have a machine with adequate memory (RAM) capacity. The required memory can be calculated using the following formula:
RAM Needed = (6 bytes Cα + 1 3Di byte + 1 AA byte) * (residues in the database).
For example, for the AFDB50 dataset, the memory requirement can be calculated as:
8 byte * 54*10^6 (Seqs) * 350 (avg. protein length) = 151G
If searching with 3Di/AA without using the --sort-by-structure-bits 0 option, the Cα information can be disregarded. This would reduce the memory requirement for the AFDB50 dataset to:
2 bytes * 54 x 10^6 (sequences) * 350 (average protein length) = 35 GB
Please note that disabling the --sort-by-structure-bits 0 option affects the final score and ranking of hits, but not the E-values themselves. Ranking alterations primarily occur for E-values less than 10^-1.
Foldseek can be installed for Linux or macOS
(1) downloading a statically compiled version For Linux computer with supports AVX2 use:
wget https://mmseqs.com/foldseek/foldseek-linux-avx2.tar.gz
tar xvzf foldseek-linux-avx2.tar.gz
export PATH=$(pwd)/foldseek/bin/:$PATH
Linux with SSE4.1
wget https://mmseqs.com/foldseek/foldseek-linux-sse41.tar.gz
tar xvzf foldseek-linux-sse41.tar.gz
export PATH=$(pwd)/foldseek/bin/:$PATH
macOS build (universal binary with SSE4.1/AVX2/M1 NEON)
wget https://mmseqs.com/foldseek/foldseek-osx-universal.tar.gz
tar xvzf foldseek-osx-universal.tar.gz
export PATH=$(pwd)/foldseek/bin/:$PATH
(2) using bioconda
conda install -c conda-forge -c bioconda foldseek
(3) compiling the from source (see below),
Compiling Foldseek from source has the advantage that it will be optimized to the specific system, which should improve its performance. To compile Foldseek git, g++ (4.9 or higher) and cmake (2.8.12 or higher) are needed. Afterwards, the foldseek binary will be located in build/bin/.
git clone https://github.com/steineggerlab/foldseek.git
cd foldseek
mkdir build
cd build
cmake -DCMAKE_BUILD_TYPE=RELEASE -DCMAKE_INSTALL_PREFIX=. ..
make
make install
export PATH=$(pwd)/bin/:$PATH
See the Customizing compilation through CMake section if you compile Foldseek on a different system than the one where it will eventually reun.
To compile Foldseek with (Apple-)Clang you need to install either XCode or the Command Line Tools.
You also need libomp. We recommend installing it using Homebrew:
brew install cmake libomp zlib bzip2
CMake currently does not correctly identify paths to libomp. Use the script in util/build_osx.sh to compile Foldseek.
The resulting binary will be placed in OUTPUT_DIR/mmseqs.
./util/build_osx.sh PATH_TO_FOLDSEEK_REPO OUTPUT_DIR
Please install the following packages with Homebrew:
brew install cmake gcc@11 zlib bzip2
Use the following cmake call:
CC="gcc-11" CXX="g++-11" cmake -DCMAKE_BUILD_TYPE=RELEASE -DCMAKE_INSTALL_PREFIX=. ..
Most of the MMseqs2 CMake options also apply to Foldseek, refer to MMseqs2's user guide for details.
Install the google-cloud-cpp package from vcpkg:
git clone https://github.com/microsoft/vcpkg.git
./vcpkg/bootstrap-vcpkg.sh
./vcpkg/vcpkg install google-cloud-cpp
Foldseek can now be compiled with GCS support:
cd path-to-foldseek
mkdir build && cd build
cmake -DHAVE_GCS=1 -DCMAKE_TOOLCHAIN_FILE=[path to vcpkg]/scripts/buildsystems/vcpkg.cmake ..
make -j $(nproc --all)
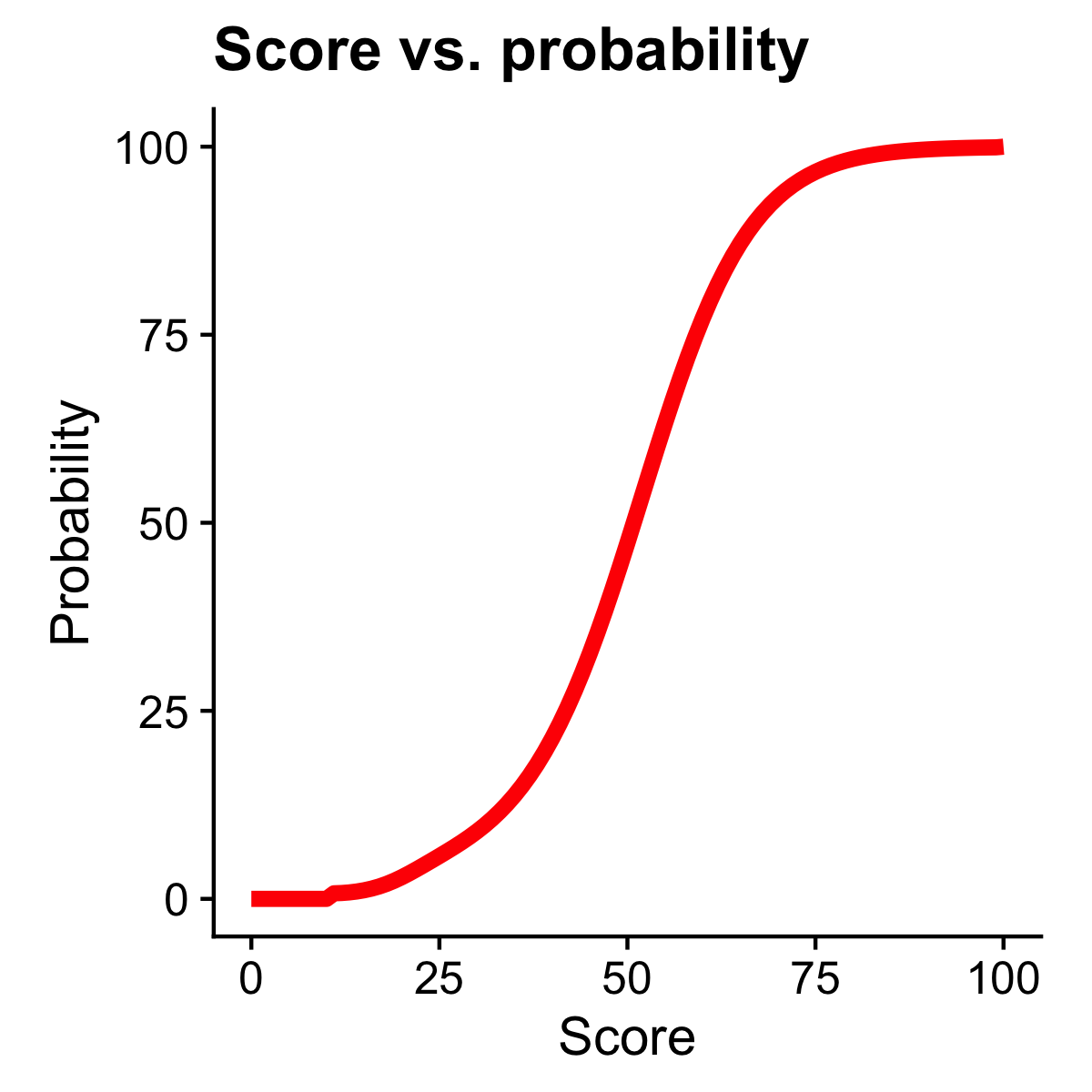
Foldseek computes for each match a simple estimate for the probability that the match is a true positive match given its structural bit score. Here, hits within the same superfamily are TP, hits to another fold are FP, and hits to the same family or to another superfamily are ignored. The probability is the fraction of TP hits from TP and FP hits found at the score on average. For this, we estimate the bit score distributions of TP and FP hits. Both score distributions were fitted on SCOPe40. For example, Foldseek finds around the same number of FP and TP with a score of 51 in SCOPe40. The probability for a hit with score 51 is therefore 50%.
In Foldseek we apply U and T to the target to superposition it onto the query structure.
Following is some awk one-liner snippet that applies the rotations to an input PDB file.
You have to provide UT as a vector of 12 values, 9 being the U matrix and 3 the T vector.
awk -v UT="-0.672446,0.740134,-0.004138,-0.740140,-0.672409,0.007633,0.002867,0.008196,0.999962,0.099348,-0.326414,-57.755688" 'BEGIN {split(UT, arr, ",")} {
if ($0 ~ /^ATOM|^HETATM/) {
x = $7
y = $8
z = $9
x_new = (x * arr[1] + y * arr[2] + z * arr[3]) + arr[10]
y_new = (x * arr[4] + y * arr[5] + z * arr[6]) + arr[11]
z_new = (x * arr[7] + y * arr[8] + z * arr[9]) + arr[12]
printf "%-6s%5d %-4s%3s %s%4d %8.3f%8.3f%8.3f%6.2f%6.2f %2s\n", $1, $2, $3, $4, $5, $6, x_new, y_new, z_new, $11, $12, $13
}
else {
print $0
}
}' input.pdb > output.pdb
You can make a your own prefiltering database to tell the structurealign module what pairs to align.
# assuming you have a query and target database
foldseek createdb inputs1/ db1
foldseek createdb inputs2/ db2
# make a mapping of the accession that you want to align (check 2nd column in the dbN.lookup file)
echo -e "d1asha_\td1b0ba_\nd1asha_\td1cg5a_\n" > to_align.tsv
# convert this into the internal numeric database keys
awk 'FNR == 1 { findex++; } \
findex == 1 { f1[$2] = $1; next; } \
findex == 2 { f2[$2] = $1; next; } \
$1 in f1 && $2 in f2 { print f1[$1]"\t"f2[$2]; }' \
db1.lookup db2.lookup <(sort -s -k1,1n to_align.tsv ) > keys.tsv
# make a fake prefiltering database
foldseek tsv2db keys.tsv pref --output-dbtype 7
# foldseek alignment
foldseek structurealign db1 db2 pref aln
# m8 human readable output
foldseek convertalis db1 db2 aln aln.m8