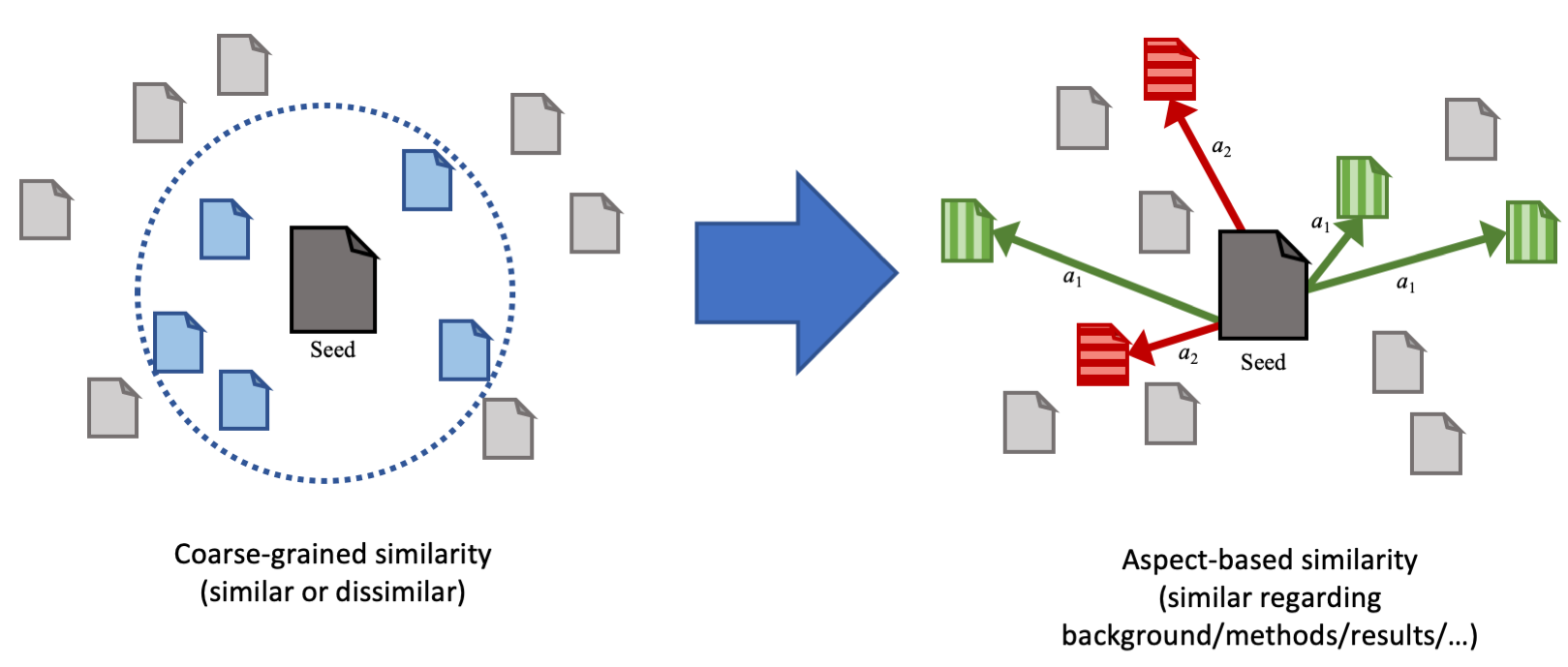
Implementation, trained models and result data for the paper Aspect-based Document Similarity for Research Papers (PDF on Arxiv). The supplemental material is available for download under GitHub Releases or Zenodo.
- Datasets are compatible with 🤗 Huggingface NLP library (now known as datasets).
- Models are available on 🤗 Huggingface Transformers models.
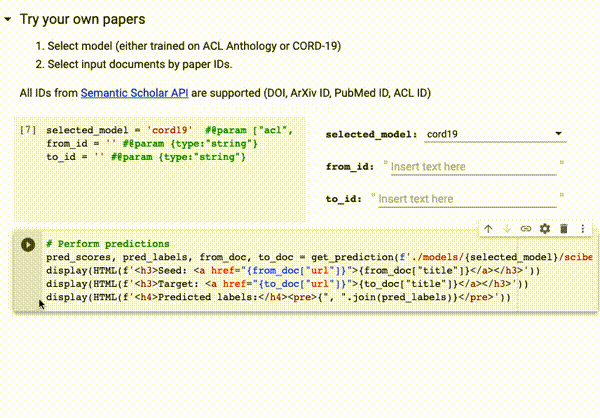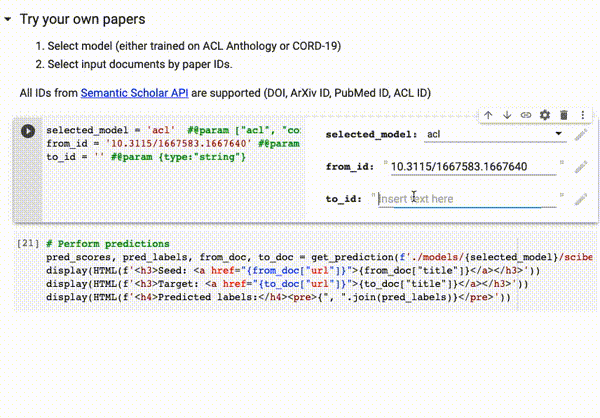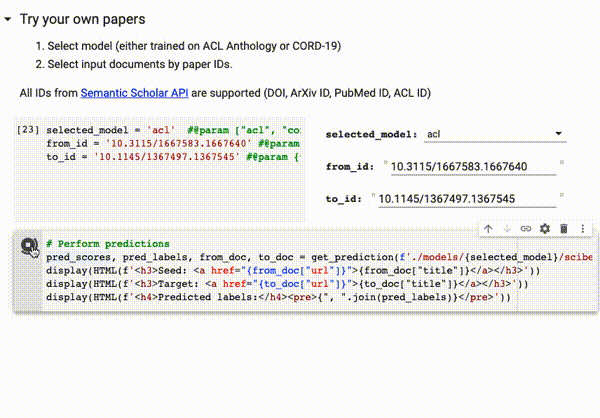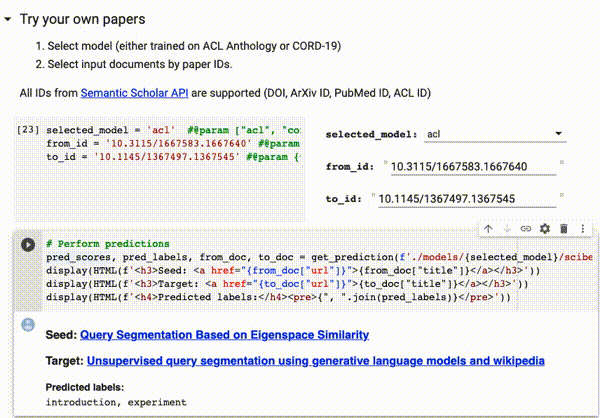
You can try our trained models directly on Google Colab on all papers available on Semantic Scholar (via DOI, ArXiv ID, ACL ID, PubMed ID):
- Python 3.7
- CUDA GPU (for Transformers)
Datasets
Create a new virtual environment for Python 3.7 with Conda:
conda create -n paper python=3.7
conda activate paperClone repository and install dependencies:
git clone https://github.com/malteos/aspect-document-similarity.git repo
cd repo
pip install -r requirements.txtTo reproduce our experiments, follow these steps (if you just want to train and test the models, skip the first two steps):
export DIR=./output
# ACL Anthology
# Get parscit files from: https://acl-arc.comp.nus.edu.sg/archives/acl-arc-160301-parscit/)
sh ./sbin/download_parsecit.sh
# CORD-19
wget https://ai2-semanticscholar-cord-19.s3-us-west-2.amazonaws.com/historical_releases/cord-19_2020-03-13.tar.gz
# Get additional data (collected from Semantic Scholar API)
wget https://github.com/malteos/aspect-document-similarity/releases/download/1.0/acl_s2.tar
wget https://github.com/malteos/aspect-document-similarity/releases/download/1.0/cord19_s2.tar# ACL
python -m acl.dataset save_dataset <input_dir> <parscit_dir> <output_dir>
# CORD-19
python -m cord19.dataset save_dataset <input_dir> <output_dir>
The datasets are built on the Huggingface NLP library (soon available on the official repository):
from nlp import load_dataset
# Training data for first CV split
train_dataset = load_dataset(
'./datasets/cord19_docrel/cord19_docrel.py',
name='relations',
split='fold_1_train'
) from models.auto_modelling import AutoModelForMultiLabelSequenceClassification
# Load models with pretrained weights from Huggingface model hub
acl_model = AutoModelForMultiLabelSequenceClassification('malteos/aspect-acl-scibert-scivocab-uncased')
cord19_model = AutoModelForMultiLabelSequenceClassification('malteos/aspect-cord19-scibert-scivocab-uncased')
# Use the models in standard Huggingface fashion ...
# acl_model(input_ids, token_type_ids, ...)
# cord19_model(input_ids, token_type_ids, ...)All models are trained with the trainer_cli.py script:
python trainer_cli.py --cv_fold $CV_FOLD \
--output_dir $OUTPUT_DIR \
--model_name_or_path $MODEL_NAME \
--doc_id_col $DOC_ID_COL \
--doc_a_col $DOC_A_COL \
--doc_b_col $DOC_B_COL \
--nlp_dataset $NLP_DATASET \
--nlp_cache_dir $NLP_CACHE_DIR \
--cache_dir $CACHE_DIR \
--num_train_epochs $EPOCHS \
--seed $SEED \
--per_gpu_eval_batch_size $EVAL_BATCH_SIZE \
--per_gpu_train_batch_size $TRAIN_BATCH_SIZE \
--learning_rate $LR \
--do_train \
--save_predictionsThe exact parameters are available in sbin/acl and sbin/cord19.
The results can be computed and viewed with a Jupyter notebook. Figures and tables from the paper are part of the notebook.
jupyter notebook evaluation.ipynbDue to the space constraints some results could not be included in the paper.
The full results for all methods and all test samples are available as
CSV files under Releases
(or via the Jupyter notebook).
If you are using our code, please cite our paper:
@InProceedings{Ostendorff2020c,
title = {Aspect-based Document Similarity for Research Papers},
booktitle = {Proceedings of the 28th International Conference on Computational Linguistics (COLING 2020)},
author = {Ostendorff, Malte and Ruas, Terry and Blume, Till and Gipp, Bela and Rehm, Georg},
year = {2020},
month = {Dec.},
}MIT