$ git clone [email protected]:cxhernandez/nma.git && cd nma
$ python setup.py install# Imports
import mdtraj as md
from nma import ANMA
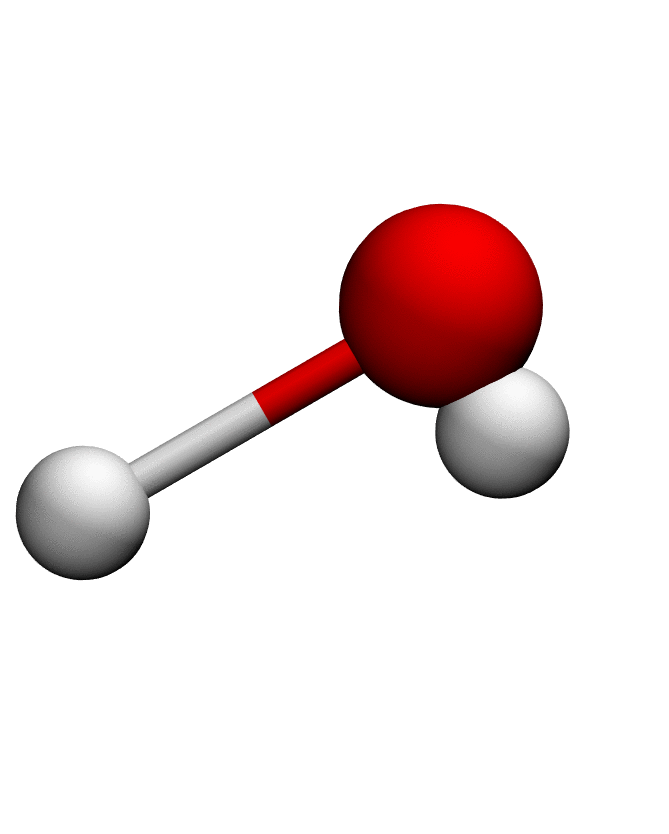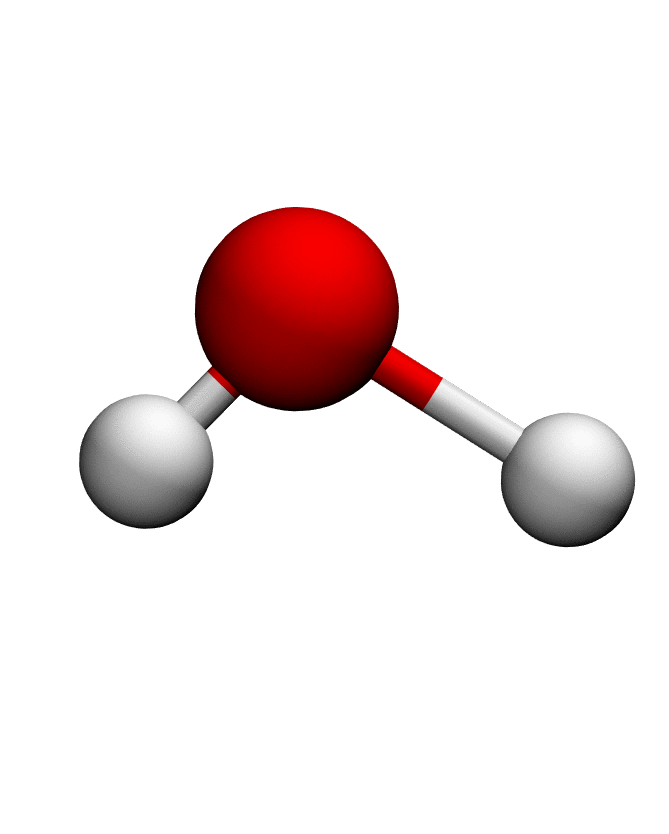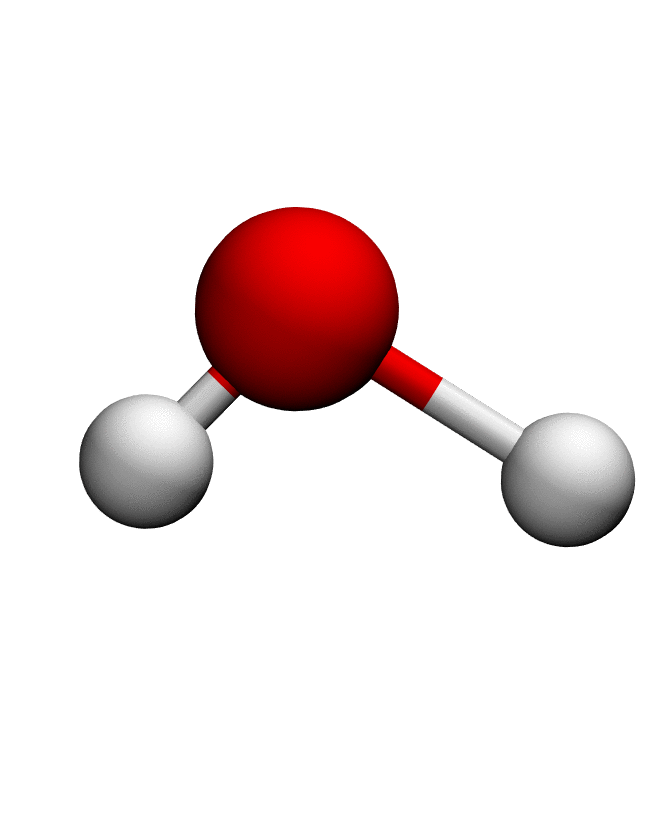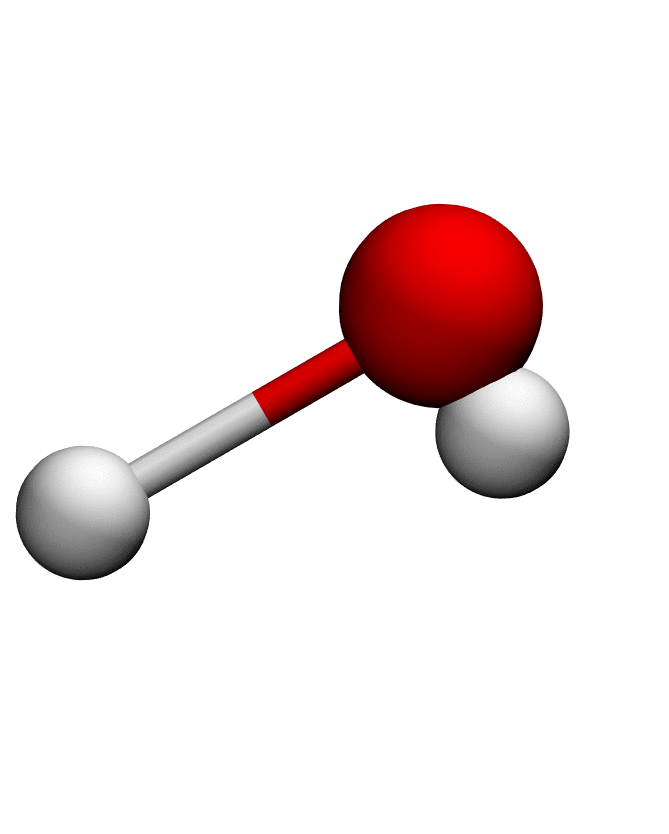
# Load structure of choice (e.g. Water)
pdb = md.load_pdb('./examples/water.pdb')
# Initialize ANMA object
anma = ANMA(mode=0, rmsd=0.06, n_steps=50, selection='all')
# Transform the PDB into a short trajectory of a given mode
anma_traj = anma.fit_transform(pdb)| Mode 0 | Mode 1 | Mode 2 |
|---|---|---|
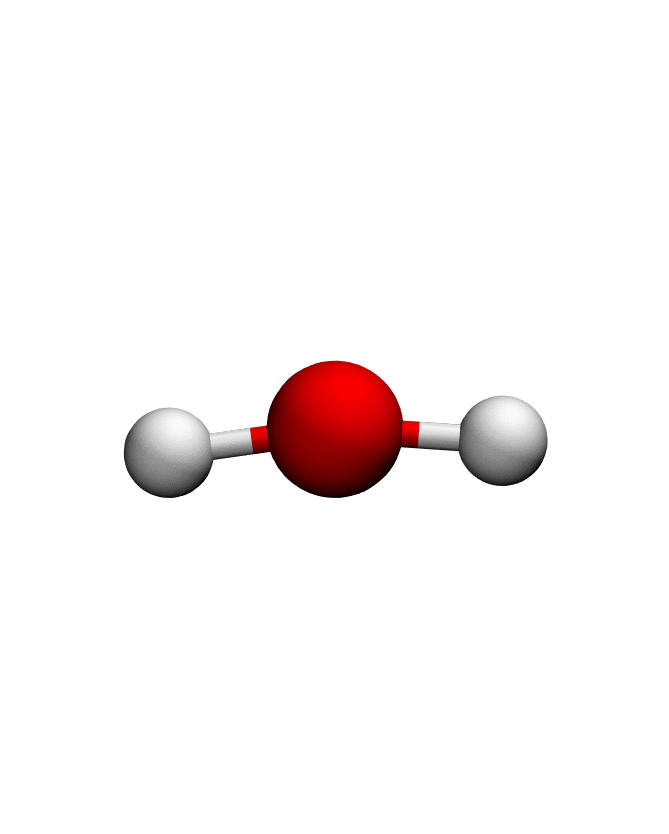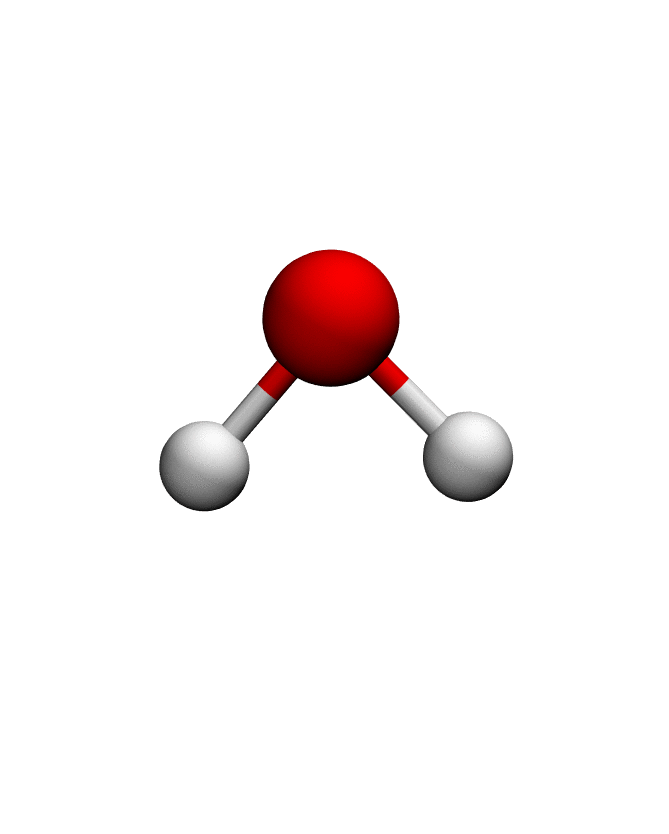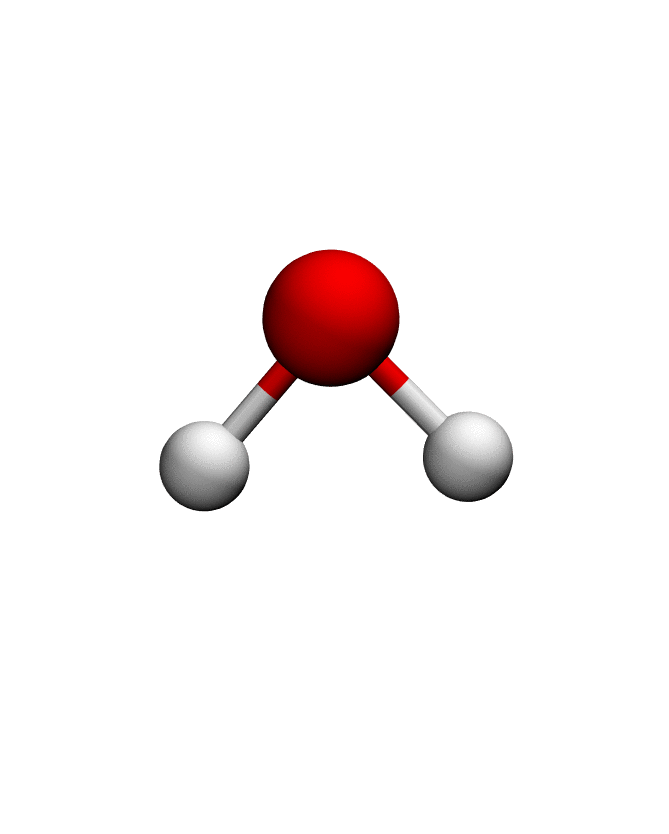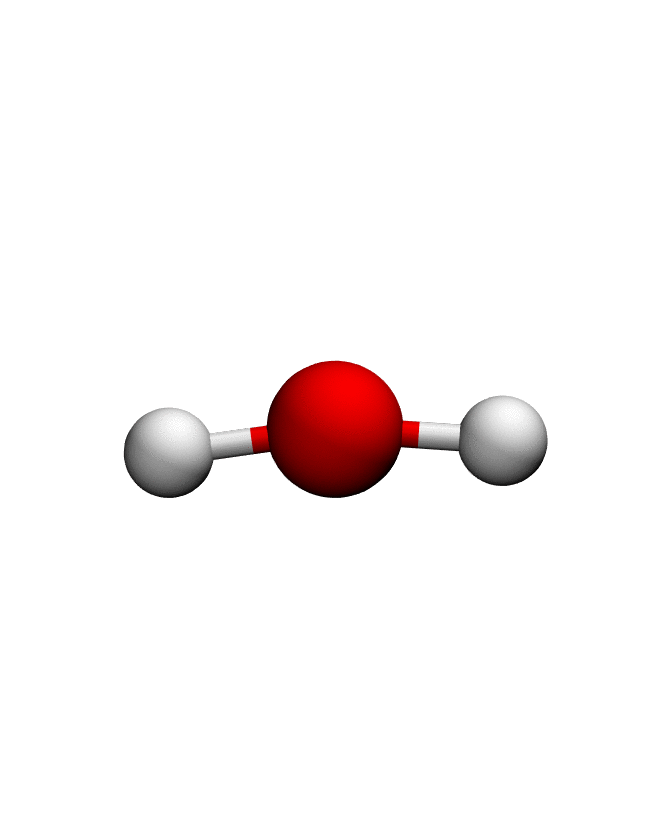 |
 |
 |
- ProDy (for loose inspiration)