-
Notifications
You must be signed in to change notification settings - Fork 619
Differential Splicing
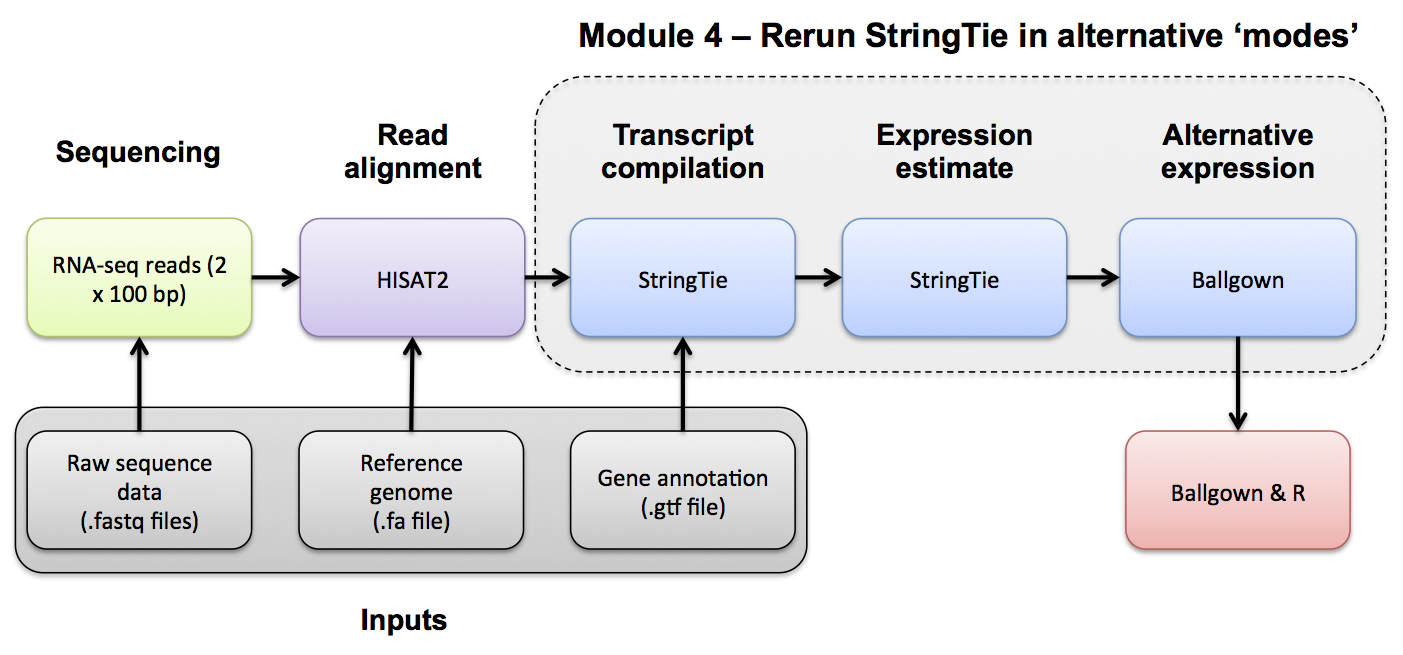
Use Ballgown and Stringtie to compare the UHR and HBR conditions against reference guided and de novo transcript assemblies.
Refer to the Stringtie manual for a more detailed explanation: https://ccb.jhu.edu/software/stringtie/index.shtml?t=manual
The Ballgown github page also has documentation for getting started with ballgown: https://github.com/alyssafrazee/ballgown
Calculate UHR and HBR expression estimates, for known/novel (reference guided mode) transcripts
Re-run Stringtie using the reference guided merged GTF, and output tables for Ballgown. Store the results in a new directory so that we can still examine the results generated without the merged GTF.
cd $RNA_HOME/expression/stringtie/
mkdir ref_guided_merged
cd ref_guided_merged
stringtie -p 8 -G ../ref_guided/stringtie_merged.gtf -e -B -o HBR_Rep1/transcripts.gtf $RNA_ALIGN_DIR/HBR_Rep1.bam
stringtie -p 8 -G ../ref_guided/stringtie_merged.gtf -e -B -o HBR_Rep2/transcripts.gtf $RNA_ALIGN_DIR/HBR_Rep2.bam
stringtie -p 8 -G ../ref_guided/stringtie_merged.gtf -e -B -o HBR_Rep3/transcripts.gtf $RNA_ALIGN_DIR/HBR_Rep3.bam
stringtie -p 8 -G ../ref_guided/stringtie_merged.gtf -e -B -o UHR_Rep1/transcripts.gtf $RNA_ALIGN_DIR/UHR_Rep1.bam
stringtie -p 8 -G ../ref_guided/stringtie_merged.gtf -e -B -o UHR_Rep2/transcripts.gtf $RNA_ALIGN_DIR/UHR_Rep2.bam
stringtie -p 8 -G ../ref_guided/stringtie_merged.gtf -e -B -o UHR_Rep3/transcripts.gtf $RNA_ALIGN_DIR/UHR_Rep3.bam
Run Ballgown using the reference guided, merged transcripts
mkdir -p $RNA_HOME/de/ballgown/ref_guided_merged/
cd $RNA_HOME/de/ballgown/ref_guided_merged/
printf "\"ids\",\"type\",\"path\"\n\"UHR_Rep1\",\"UHR\",\"$RNA_HOME/expression/stringtie/ref_guided_merged/UHR_Rep1\"\n\"UHR_Rep2\",\"UHR\",\"$RNA_HOME/expression/stringtie/ref_guided_merged/UHR_Rep2\"\n\"UHR_Rep3\",\"UHR\",\"$RNA_HOME/expression/stringtie/ref_guided_merged/UHR_Rep3\"\n\"HBR_Rep1\",\"HBR\",\"$RNA_HOME/expression/stringtie/ref_guided_merged/HBR_Rep1\"\n\"HBR_Rep2\",\"HBR\",\"$RNA_HOME/expression/stringtie/ref_guided_merged/HBR_Rep2\"\n\"HBR_Rep3\",\"HBR\",\"$RNA_HOME/expression/stringtie/ref_guided_merged/HBR_Rep3\"\n" > UHR_vs_HBR.csv
Please see Differential Expression for details on running ballgown to determine a DE gene/transcript list.
Calculate UHR and HBR expression estimates, for known/novel (de novo mode) transcripts:
Re-run Stringtie using the de novo merged GTF, and output tables for Ballgown. Store the results in a new directory so that we can still examine the results generated without the merged GTF.
cd $RNA_HOME/expression/stringtie/de_novo
cd $RNA_HOME/expression/stringtie/
mkdir de_novo_merged
cd de_novo_merged
stringtie -p 8 -G ../de_novo/stringtie_merged.gtf -e -B -o HBR_Rep1/transcripts.gtf $RNA_ALIGN_DIR/HBR_Rep1.bam
stringtie -p 8 -G ../de_novo/stringtie_merged.gtf -e -B -o HBR_Rep2/transcripts.gtf $RNA_ALIGN_DIR/HBR_Rep2.bam
stringtie -p 8 -G ../de_novo/stringtie_merged.gtf -e -B -o HBR_Rep3/transcripts.gtf $RNA_ALIGN_DIR/HBR_Rep3.bam
stringtie -p 8 -G ../de_novo/stringtie_merged.gtf -e -B -o UHR_Rep1/transcripts.gtf $RNA_ALIGN_DIR/UHR_Rep1.bam
stringtie -p 8 -G ../de_novo/stringtie_merged.gtf -e -B -o UHR_Rep2/transcripts.gtf $RNA_ALIGN_DIR/UHR_Rep2.bam
stringtie -p 8 -G ../de_novo/stringtie_merged.gtf -e -B -o UHR_Rep3/transcripts.gtf $RNA_ALIGN_DIR/UHR_Rep3.bam
Run Ballgown using the de novo, merged transcripts
mkdir -p $RNA_HOME/de/ballgown/de_novo_merged/
cd $RNA_HOME/de/ballgown/de_novo_merged/
printf "\"ids\",\"type\",\"path\"\n\"UHR_Rep1\",\"UHR\",\"$RNA_HOME/expression/stringtie/de_novo_merged/UHR_Rep1\"\n\"UHR_Rep2\",\"UHR\",\"$RNA_HOME/expression/stringtie/de_novo_merged/UHR_Rep2\"\n\"UHR_Rep3\",\"UHR\",\"$RNA_HOME/expression/stringtie/de_novo_merged/UHR_Rep3\"\n\"HBR_Rep1\",\"HBR\",\"$RNA_HOME/expression/stringtie/de_novo_merged/HBR_Rep1\"\n\"HBR_Rep2\",\"HBR\",\"$RNA_HOME/expression/stringtie/de_novo_merged/HBR_Rep2\"\n\"HBR_Rep3\",\"HBR\",\"$RNA_HOME/expression/stringtie/de_novo_merged/HBR_Rep3\"\n" > UHR_vs_HBR.csv
Please see Differential Expression for details on running ballgown to determine a DE gene/transcript list.
| Previous Section | This Section | Next Section |
|---|---|---|
| Merging | Differential Splicing | Splicing Visualization |
NOTICE: This resource has been moved to rnabio.org. The version here will be maintained for legacy use only. All future development and maintenance will occur only at rnabio.org. Please proceed to rnabio.org for the current version of this course.
Table of Contents
Module 0: Authors | Citation | Syntax | Intro to AWS | Log into AWS | Unix | Environment | Resources
Module 1: Installation | Reference Genomes | Annotations | Indexing | Data | Data QC
Module 2: Adapter Trim | Alignment | IGV | Alignment Visualization | Alignment QC
Module 3: Expression | Differential Expression | DE Visualization
Module 4: Alignment Free - Kallisto
Module 5: Ref Guided | De novo | Merging | Differential Splicing | Splicing Visualization
Module 6: Trinity
Module 7: Trinotate
Appendix: Saving Results | Abbreviations | Lectures | Practical Exercise Solutions | Integrated Assignment | Proposed Improvements | AWS Setup