-
Notifications
You must be signed in to change notification settings - Fork 1
Coexpression Analysis
Multiple methods are implemented in MOG to find correlations and associations among features or samples.
All methods statistical methods are accessed by CLICKing 
Statistical analysis button in the menubar of the "Feature metadata" pane.
The "Analyze" button in the menubar of the "Sample Metadata Table pane" displays the options available to calculate statistical associations between and among samples.
Coexpression methods implemented in MOG:
Pearson Correlation detects the linear dependency between two variables X and Y.
Spearman Correlation coefficient measures monotonic relationships between two variables X and Y.
Mutual Information (MI) quantifies the amount of information shared between two random variables. MI for two discrete random variables X and Y, having the joint probability p(x,y) and marginal probabilities p(x) and p(y) respectively, is defined as:

Correlations between features are helpful in finding features that are coexpressed across samples. To calculate pairwise correlations of a selected feature with all the other features in the list:
-
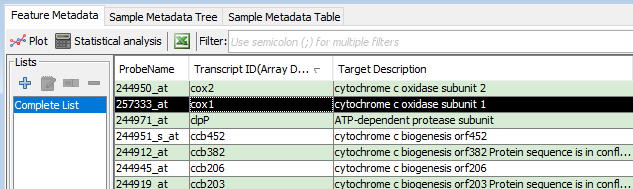
In the "Feature metadata" pane., select a single row to calculate the correlation against.
-

CLICK the "Statistical Analysis" button in the "Feature metadata" pane. menubar. -

Select a coexpression method (Pearson, Spearman, or MI). When prompted, enter a name to save your results. This name will be saved in the "Feature Metadata" table. -
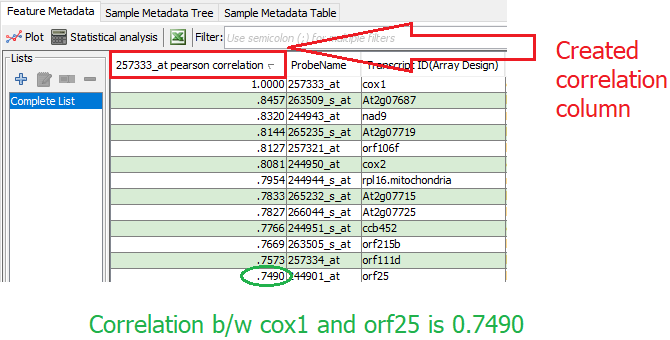
The i^{th} row in the column represents the computed correlation value of selected feature with the i^{th} feature.
The column containing the coexpression results is saved with the MOG project. Correlation columns can be deleted by selecting "Remove Correlation" in step 2. The "View correlation details" option in step 2 shows the details (if available) about the computed correlation. For example, if a correlation is computed with a P-Value then the p-values could be seen with "View correlation details". To remove a MOG-computed correlation:
-

CLICK the "Statistical Analysis" button in the "Feature Metadata" pane menubar. Select "Remove Correlation". This will display a list of saved correlations. Select the ones which you want removed.
MOG applies a permutation test to compute the p-values for correlations. MOG uses multithreading to speed-up this intensive computation, processing each permuted dataset in parallel (Algorithm 1). MOG provides Bonferroni corrections and Benjamini--Hochberg (BH) corrections to adjust the p-values for multiple comparisons.

For permutation (8 (Algorithm 1)) the user can select blocks to permute the data, such that the data in the blocks are exchangeable under the null hypothesis. The blocks are determined by the experimental design. For, example if the metadata is structured in the tree of Figure 27, the user may choose to permute data only within samples. MOG keeps a variable called "Replicate column" which stores a chosen factor, from the sample metadata, by the user (see Section 5.1).

Figure 12: Selecting Pearson correlation matrix
Pairwise matrices of correlations of each feature to each other feature serve as the basis for clustering. This has been implemented for Pearson and Spearman correlations.
- To compute Pearson Correlation matrix: Select "Differential correlation" option through "Statistical Analysis --> Correlation --> Pearson correlation matrix (see Figure 12).
- To compute Pearson Correlation matrix: Select "Differential correlation" option through "Statistical Analysis --> Correlation --> Spearman correlation matrix.
- Enter file name to save the results.
Note: Computing all pairwise correlations for thousands of features can take significant time (minutes) depending on the number of samples. To compute pairwise correlations for only a given set of features, make a list of those features and select that list before the analysis (see Feature List).

Figure 13 Selecting differential correlation.
MOG can test whether features are differentially correlated between two groups using the Fisher transformation method. Differential correlations means that the patterns of coexpression in a group of samples differ from that in a different group of samples. Features with differential correlation may reveal changes in biological interactions under different sets of conditions. To compute differential correlation for a given feature under between two groups of samples:
- Select "Differential correlation" option through "Statistical Analysis --> Correlation --> Differential correlation --> New Differential Correlation (see Figure 13).
- A dialog identical to "Differential Expression Analysis" window will appear.
- Add samples to the groups (see "Differential Expression Analysis" window)
- CLICK OK. The results will be displayed in a new window.
To compute differential correlation from existing Pearson's correlation columns:
- Select "Differential correlation" option through "Statistical Analysis --> Correlation --> Differential correlation --> From Existing Columns (see Figure 13).
- Select the first correlation column and enter N1 which is the number of samples used in the calculation.
- Select the second correlation column and enter N2 which is the number of samples used in the calculation.
- The results will be displayed in a new window.
Note: Because the goal of finding differential correlation is to identify if the correlations of a feature with other features changes over different groups, the user must carefully choose the correlation columns (which contains the correlation of a feature with other features under different samples) and enter the correct group sizes. The interpretation of the results is up to the user.
Correlation between samples can reveal how similar various different samples are to one another other. This can be helpful in revealing the similarity between samples from different biological groups and evaluating the importance of technical effects. It also can determine whether a set of "replicates" are similar or very diverse To find the correlation between selected samples:
-
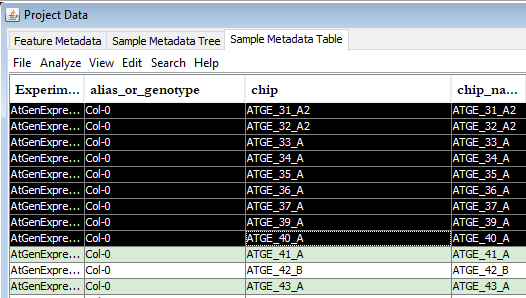
In the "Sample Metadata Table pane", select the required rows (each row represents a sample). -

In the "Sample Metadata Table pane" menubar, select the "Analyze" option and choose the appropriate method. -

The results are displayed in a new window. The results could be saved to file by going to File --> Export.
2. THE BASICS: Downloading and Using MOG
5. Sort, subset, transform, analyze, and reorder the Data
8. Differential Expression Analysis
12. Change Project Properties and MOG Properties
2.3 Download a pre-compiled project
2.5 Run MetaOmGraph by providing more memory(RAM)
5.1 Choosing the Replicate Column
5.5 Search or Filter Sample MetaData
6.1 Correlation Between Features
6.3 Correlation Betweeen Samples
7.2 Line Charts with Averaged Replicates
8.1 Differential Expression Analysis Window
8.2 Differential Expression Results Window
9.1 Principal component analysis (PCA)