InDEP: an interpretable machine learning approach to predict cancer driver genes from multi-omics data
It's my first study about interpreting the model of predicting cancer driver genes. The study proposed a machine learning framework named named InDEP. ^ ^
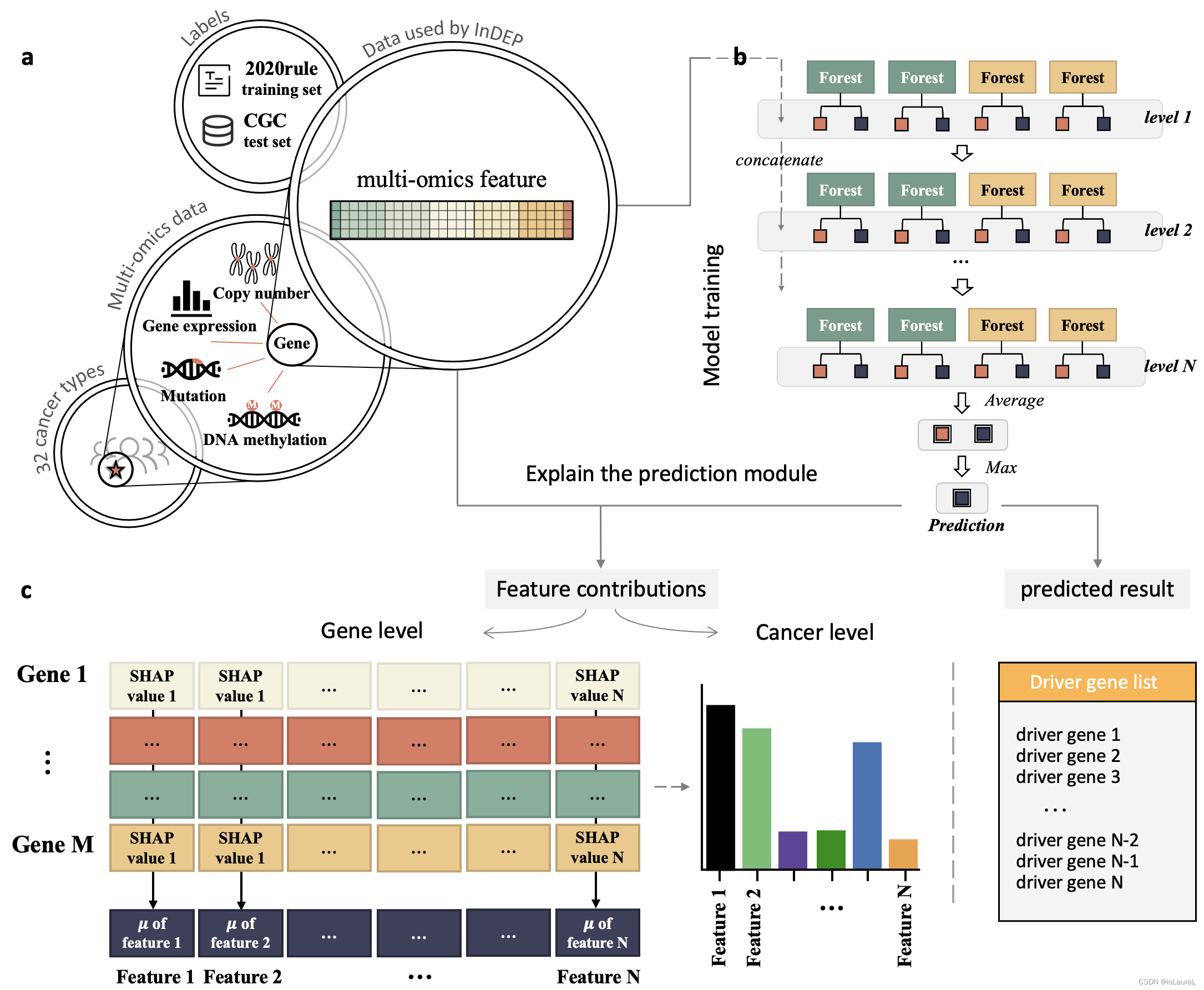
This study aims to address this issue by introducing InDEP, an interpretable machine learning framework based on cascade forests. InDEP is designed with easy-to-interpret features, cascade forests based on decision trees, and a KernelSHAP module that enables fine-grained post-hoc interpretation. Integrating multiomics data, InDEP can identify essential features of classified driver genes at both the gene and cancer-type levels. The framework accurately identifies driver genes, discovers new patterns that make genes as driver genes from the prediction process, and refines the cancer driver gene catalog. In comparison to state-of-the-art methods, InDEP proved to be more accurate on the test set and identified reliable candidate driver genes. Mutational features were the primary drivers for InDEP’s identifying driver genes, with other omics features also contributing. At the gene level, the framework concluded that substitution-type mutations were the main reason most genes were identified as driver genes.
The study is awaiting reviewer's scores in briefings in bioinformatics... Good Luck~!
------中国时间2023.8.17 英国时间2023.8.16-------
你怎么知道笔者的论文在今天收录啦~ 哈哈~ 毕业有望~
cd InDEP
pip install -r requirement.txt
cd codes
for example:
train the model
python run.py -t PANCAN -m train -l InDEP
get the score of genes
python run.py -t PANCAN -m score -l InDEP
test the model
python run.py -t PANCAN -m eval -l InDEP
get the interpretation of the model
python run.py -t PANCAN -m interpretation -l InDEP
Now, it supports more features, you can use "-r origin,noncoding,cnv,CADD,subtype,norm_exp" to choose what you want to use~~