-
Notifications
You must be signed in to change notification settings - Fork 6
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
16 changed files
with
11,143 additions
and
1,113 deletions.
There are no files selected for viewing
4 changes: 2 additions & 2 deletions
4
_freeze/schedule/slides/09-l1-penalties/execute-results/html.json
Large diffs are not rendered by default.
Oops, something went wrong.
18 changes: 18 additions & 0 deletions
18
_freeze/schedule/slides/12-why-smooth/execute-results/html.json
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,18 @@ | ||
| { | ||
| "hash": "3c5683f23b8b76620620eb2b5397af37", | ||
| "result": { | ||
| "markdown": "---\nlecture: \"12 To(o) smooth or not to(o) smooth?\"\nformat: revealjs\nmetadata-files: \n - _metadata.yml\n---\n---\n---\n\n## {{< meta lecture >}} {.large background-image=\"gfx/smooths.svg\" background-opacity=\"0.3\"}\n\n[Stat 406]{.secondary}\n\n[{{< meta author >}}]{.secondary}\n\nLast modified -- 02 October 2023\n\n\n\n$$\n\\DeclareMathOperator*{\\argmin}{argmin}\n\\DeclareMathOperator*{\\argmax}{argmax}\n\\DeclareMathOperator*{\\minimize}{minimize}\n\\DeclareMathOperator*{\\maximize}{maximize}\n\\DeclareMathOperator*{\\find}{find}\n\\DeclareMathOperator{\\st}{subject\\,\\,to}\n\\newcommand{\\E}{E}\n\\newcommand{\\Expect}[1]{\\E\\left[ #1 \\right]}\n\\newcommand{\\Var}[1]{\\mathrm{Var}\\left[ #1 \\right]}\n\\newcommand{\\Cov}[2]{\\mathrm{Cov}\\left[#1,\\ #2\\right]}\n\\newcommand{\\given}{\\ \\vert\\ }\n\\newcommand{\\X}{\\mathbf{X}}\n\\newcommand{\\x}{\\mathbf{x}}\n\\newcommand{\\y}{\\mathbf{y}}\n\\newcommand{\\P}{\\mathcal{P}}\n\\newcommand{\\R}{\\mathbb{R}}\n\\newcommand{\\norm}[1]{\\left\\lVert #1 \\right\\rVert}\n\\newcommand{\\snorm}[1]{\\lVert #1 \\rVert}\n\\newcommand{\\tr}[1]{\\mbox{tr}(#1)}\n\\newcommand{\\brt}{\\widehat{\\beta}^R_{s}}\n\\newcommand{\\brl}{\\widehat{\\beta}^R_{\\lambda}}\n\\newcommand{\\bls}{\\widehat{\\beta}_{ols}}\n\\newcommand{\\blt}{\\widehat{\\beta}^L_{s}}\n\\newcommand{\\bll}{\\widehat{\\beta}^L_{\\lambda}}\n$$\n\n\n\n\n\n## Last time...\n\n\nWe've been discussing smoothing methods in 1-dimension:\n\n$$\\Expect{Y\\given X=x} = f(x),\\quad x\\in\\R$$\n\nWe looked at basis expansions, e.g.:\n\n$$f(x) \\approx \\beta_0 + \\beta_1 x + \\beta_2 x^2 + \\cdots + \\beta_k x^k$$ \n\nWe looked at local methods, e.g.:\n\n$$f(x_i) \\approx s_i^\\top \\y$$\n\n. . .\n\nWhat if $x \\in \\R^p$ and $p>1$?\n\n::: aside\nNote that $p$ means the dimension of $x$, not the dimension of the space of the polynomial basis or something else. That's why I put $k$ above.\n:::\n\n\n\n## Kernels and interactions\n\nIn multivariate nonparametric regression, you estimate a [surface]{.secondary} over the input variables.\n\nThis is trying essentially to find $\\widehat{f}(x_1,\\ldots,x_p)$.\n\nTherefore, this function [by construction]{.secondary} includes interactions, handles categorical data, etc. etc.\n\nThis is in contrast with explicit [linear models]{.secondary} which need you to specify these things.\n\nThis extra complexity (automatically including interactions, as well as other things) comes with tradeoffs.\n\n. . . \n\nMore complicated functions (smooth Kernel regressions vs. linear models) tend to have [lower bias]{.secondary} but [higher variance]{.secondary}.\n\n\n\n## Issue 1\n\nFor $p=1$, one can show that for kernels (with the correct bandwidth)\n\n$$\\textrm{MSE}(\\hat{f}) = \\frac{C_1}{n^{4/5}} + \\frac{C_2}{n^{4/5}} + \\sigma^2$$ \n\n\n::: callout-danger\n\n_you don't need to memorize these formulas_ but you should know the intuition\n\n:::\n\n## Issue 1\n\nFor $p=1$, one can show that for kernels (with the correct bandwidth)\n\n$$\\textrm{MSE}(\\hat{f}) = \\frac{C_1}{n^{4/5}} + \\frac{C_2}{n^{4/5}} + \\sigma^2$$ \n\n\nRecall, this decomposition is **squared bias** + **variance** + **irreducible error**\n\n* It depends on the **choice** of $h$\n\n$$\\textrm{MSE}(\\hat{f}) = C_1 h^4 + \\frac{C_2}{nh} + \\sigma^2$$ \n\n* Using $h = cn^{-1/5}$ **balances** squared bias and variance, leads to the above rate. (That balance minimizes the MSE)\n\n## Issue 1\n\n\nFor $p=1$, one can show that for kernels (with the correct bandwidth)\n\n$$\\textrm{MSE}(\\hat{f}) = \\frac{C_1}{n^{4/5}} + \\frac{C_2}{n^{4/5}} + \\sigma^2$$ \n\n\n### Intuition: \n\nas you collect data, use a smaller bandwidth and the MSE (on future data) decreases\n\n\n## Issue 1\n\nFor $p=1$, one can show that for kernels (with the correct bandwidth)\n\n$$\\textrm{MSE}(\\hat{f}) = \\frac{C_1}{n^{4/5}} + \\frac{C_2}{n^{4/5}} + \\sigma^2$$ \n\n\n[How does this compare to just using a linear model?]{.primary}\n\n[Bias]{.secondary}\n \n1. The bias of using a linear model **if the truth nonlinear** is a number $b > 0$ which doesn't depend on $n$.\n2. The bias of using kernel regression is $C_1/n^{4/5}$. This goes to 0 as $n\\rightarrow\\infty$.\n \n[Variance]{.secondary}\n\n1. The variance of using a linear model is $C/n$ **no matter what**\n2. The variance of using kernel regression is $C_2/n^{4/5}$.\n\n## Issue 1\n\n\nFor $p=1$, one can show that for kernels (with the correct bandwidth)\n\n$$\\textrm{MSE}(\\hat{f}) = \\frac{C_1}{n^{4/5}} + \\frac{C_2}{n^{4/5}} + \\sigma^2$$ \n\n\n### To conclude: \n\n* bias of kernels goes to zero, bias of lines doesn't (unless the truth is linear).\n\n* but variance of lines goes to zero faster than for kernels.\n\nIf the linear model is **right**, you win. \n\nBut if it's wrong, you (eventually) lose as $n$ grows.\n\nHow do you know if you have enough data? \n\nCompare of the kernel version with CV-selected tuning parameter with the estimate of the risk for the linear model.\n\n\n# ☠️☠️ Danger ☠️☠️\n\nYou can't just compare the CVM for the kernel version to the CVM for the LM. This is because you used CVM to select the tuning parameter, so we're back to the usual problem of using the data twice. You have to do [another]{.hand} CV to estimate the risk of the kernel version at CV selected tuning parameter.\n️\n\n\n## Issue 2\n\nFor $p>1$, there is more trouble.\n\nFirst, lets look again at \n$$\\textrm{MSE}(\\hat{f}) = \\frac{C_1}{n^{4/5}} + \\frac{C_2}{n^{4/5}} + \\sigma^2$$ \n\nThat is for $p=1$. It's not __that much__ slower than $C/n$, the variance for linear models.\n\nIf $p>1$ similar calculations show,\n\n$$\\textrm{MSE}(\\hat f) = \\frac{C_1+C_2}{n^{4/(4+p)}} + \\sigma^2 \\hspace{2em} \\textrm{MSE}(\\hat \\beta) = b + \\frac{Cp}{n} + \\sigma^2 .$$\n\n## Issue 2\n\n$$\\textrm{MSE}(\\hat f) = \\frac{C_1+C_2}{n^{4/(4+p)}} + \\sigma^2 \\hspace{2em} \\textrm{MSE}(\\hat \\beta) = b + \\frac{Cp}{n} + \\sigma^2 .$$\n\n\nWhat if $p$ is big (and $n$ is really big)?\n\n1. Then $(C_1 + C_2) / n^{4/(4+p)}$ is still big.\n2. But $Cp / n$ is small.\n3. So unless $b$ is big, we should use the linear model.\n \nHow do you tell? Do model selection to decide.\n\nA [very, very]{.secondary} questionable rule of thumb: if $p>\\log(n)$, don't do smoothing.\n\n\n# Next time...\n\nCompromises if _p_ is big\n\nAdditive models and trees\n", | ||
| "supporting": [], | ||
| "filters": [ | ||
| "rmarkdown/pagebreak.lua" | ||
| ], | ||
| "includes": { | ||
| "include-after-body": [ | ||
| "\n<script>\n // htmlwidgets need to know to resize themselves when slides are shown/hidden.\n // Fire the \"slideenter\" event (handled by htmlwidgets.js) when the current\n // slide changes (different for each slide format).\n (function () {\n // dispatch for htmlwidgets\n function fireSlideEnter() {\n const event = window.document.createEvent(\"Event\");\n event.initEvent(\"slideenter\", true, true);\n window.document.dispatchEvent(event);\n }\n\n function fireSlideChanged(previousSlide, currentSlide) {\n fireSlideEnter();\n\n // dispatch for shiny\n if (window.jQuery) {\n if (previousSlide) {\n window.jQuery(previousSlide).trigger(\"hidden\");\n }\n if (currentSlide) {\n window.jQuery(currentSlide).trigger(\"shown\");\n }\n }\n }\n\n // hookup for slidy\n if (window.w3c_slidy) {\n window.w3c_slidy.add_observer(function (slide_num) {\n // slide_num starts at position 1\n fireSlideChanged(null, w3c_slidy.slides[slide_num - 1]);\n });\n }\n\n })();\n</script>\n\n" | ||
| ] | ||
| }, | ||
| "engineDependencies": {}, | ||
| "preserve": {}, | ||
| "postProcess": true | ||
| } | ||
| } |
20 changes: 20 additions & 0 deletions
20
_freeze/schedule/slides/13-gams-trees/execute-results/html.json
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,20 @@ | ||
| { | ||
| "hash": "63d6dde203bf88aa5c68f134e47c480f", | ||
| "result": { | ||
| "markdown": "---\nlecture: \"13 GAMs and Trees\"\nformat: revealjs\nmetadata-files: \n - _metadata.yml\n---\n---\n---\n\n## {{< meta lecture >}} {.large background-image=\"gfx/smooths.svg\" background-opacity=\"0.3\"}\n\n[Stat 406]{.secondary}\n\n[{{< meta author >}}]{.secondary}\n\nLast modified -- 02 October 2023\n\n\n\n$$\n\\DeclareMathOperator*{\\argmin}{argmin}\n\\DeclareMathOperator*{\\argmax}{argmax}\n\\DeclareMathOperator*{\\minimize}{minimize}\n\\DeclareMathOperator*{\\maximize}{maximize}\n\\DeclareMathOperator*{\\find}{find}\n\\DeclareMathOperator{\\st}{subject\\,\\,to}\n\\newcommand{\\E}{E}\n\\newcommand{\\Expect}[1]{\\E\\left[ #1 \\right]}\n\\newcommand{\\Var}[1]{\\mathrm{Var}\\left[ #1 \\right]}\n\\newcommand{\\Cov}[2]{\\mathrm{Cov}\\left[#1,\\ #2\\right]}\n\\newcommand{\\given}{\\ \\vert\\ }\n\\newcommand{\\X}{\\mathbf{X}}\n\\newcommand{\\x}{\\mathbf{x}}\n\\newcommand{\\y}{\\mathbf{y}}\n\\newcommand{\\P}{\\mathcal{P}}\n\\newcommand{\\R}{\\mathbb{R}}\n\\newcommand{\\norm}[1]{\\left\\lVert #1 \\right\\rVert}\n\\newcommand{\\snorm}[1]{\\lVert #1 \\rVert}\n\\newcommand{\\tr}[1]{\\mbox{tr}(#1)}\n\\newcommand{\\brt}{\\widehat{\\beta}^R_{s}}\n\\newcommand{\\brl}{\\widehat{\\beta}^R_{\\lambda}}\n\\newcommand{\\bls}{\\widehat{\\beta}_{ols}}\n\\newcommand{\\blt}{\\widehat{\\beta}^L_{s}}\n\\newcommand{\\bll}{\\widehat{\\beta}^L_{\\lambda}}\n$$\n\n\n\n\n\n## GAMs\n\nLast time we discussed smoothing in multiple dimensions.\n\n\nHere we introduce the concept of GAMs ([G]{.secondary}eneralized [A]{.secondary}dditive [M]{.secondary}odels)\n\nThe basic idea is to imagine that the response is the sum of some functions of the predictors:\n\n$$\\Expect{Y \\given X=x} = \\beta_0 + f_1(x_{1})+\\cdots+f_p(x_{p}).$$\n\n\nNote that OLS __is__ a GAM (take $f_j(x_{j})=\\beta_j x_{j}$):\n\n$$\\Expect{Y \\given X=x} = \\beta_0 + \\beta_1 x_{1}+\\cdots+\\beta_p x_{p}.$$\n\n## Gams\n\nThese work by estimating each $f_i$ using basis expansions in predictor $i$\n\nThe algorithm for fitting these things is called \"backfitting\":\n\n\n1. Center $\\y$ and $\\X$.\n2. Hold $f_k$ for all $k\\neq j$ fixed, and regress $f_j$ on the partial residuals using your favorite smoother.\n3. Repeat for $1\\leq j\\leq p$.\n4. Repeat steps 2 and 3 until the estimated functions \"stop moving\" (iterate)\n5. Return the results.\n\n\n\n## Very small example\n\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code}\nlibrary(mgcv)\nset.seed(12345)\nn <- 500\nsimple <- tibble(\n x1 = runif(n, 0, 2*pi),\n x2 = runif(n),\n y = 5 + 2 * sin(x1) + 8 * sqrt(x2) + rnorm(n, sd = .25)\n)\n\npivot_longer(simple, -y, names_to = \"predictor\", values_to = \"x\") |>\n ggplot(aes(x, y)) +\n geom_point(col = blue) +\n facet_wrap(~predictor, scales = \"free_x\")\n```\n\n::: {.cell-output-display}\n{fig-align='center'}\n:::\n:::\n\n\n## Very small example\n\nSmooth each coordinate independently\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code}\nex_smooth <- gam(y ~ s(x1) + s(x2), data = simple)\n# s(z) means \"smooth\" z, uses spline basis for each with ridge penalty, GCV\nplot(ex_smooth, pages = 1, scale = 0, shade = TRUE, \n resid = TRUE, se = 2, las = 1)\n```\n\n::: {.cell-output-display}\n{fig-align='center'}\n:::\n\n```{.r .cell-code}\nhead(coef(ex_smooth))\n```\n\n::: {.cell-output .cell-output-stdout}\n```\n(Intercept) s(x1).1 s(x1).2 s(x1).3 s(x1).4 s(x1).5 \n 10.2070490 -4.5764100 0.7117161 0.4548928 0.5535001 -0.2092996 \n```\n:::\n\n```{.r .cell-code}\nex_smooth$gcv.ubre\n```\n\n::: {.cell-output .cell-output-stdout}\n```\n GCV.Cp \n0.06619721 \n```\n:::\n:::\n\n\n\n## Wherefore GAMs?\n\n\nIf \n\n$\\Expect{Y \\given X=x} = \\beta_0 + f_1(x_{1})+\\cdots+f_p(x_{p}),$\n\nthen\n\n$\\textrm{MSE}(\\hat f) = \\frac{Cp}{n^{4/5}} + \\sigma^2.$\n\n* Exponent no longer depends on $p$. Converges faster. (If the truth is additive.)\n\n* You could also use the same methods to include \"some\" interactions like\n\n$$\\begin{aligned}&\\Expect{Y \\given X=x}\\\\ &= \\beta_0 + f_{12}(x_{1},\\ x_{2})+f_3(x_3)+\\cdots+f_p(x_{p}),\\end{aligned}$$\n\n## Very small example\n\nSmooth two coordinates together\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code}\nex_smooth2 <- gam(y ~ s(x1, x2), data = simple)\nplot(ex_smooth2,\n scheme = 2, scale = 0, shade = TRUE,\n resid = TRUE, se = 2, las = 1\n)\n```\n\n::: {.cell-output-display}\n{fig-align='center'}\n:::\n:::\n\n\n\n\n## Regression trees\n\nTrees involve stratifying or segmenting the predictor space into a number of simple regions.\n\nTrees are simple and useful for interpretation. \n\nBasic trees are not great at prediction. \n\nModern methods that use trees are much better (Module 4)\n\n## Regression trees\n\nRegression trees estimate piece-wise constant functions\n\nThe slabs are axis-parallel rectangles $R_1,\\ldots,R_K$ based on $\\X$\n\nIn each region, we average the $y_i$'s: $\\hat\\mu_1,\\ldots,\\hat\\mu_k$\n\nMinimize $\\sum_{k=1}^K \\sum_{i=1}^n (y_i-\\mu_k)^2$ over $R_k,\\mu_k$ for $k\\in \\{1,\\ldots,K\\}$\n\n. . .\n\nThis sounds more complicated than it is.\n\nThe minimization is performed __greedily__ (like forward stepwise regression).\n\n\n\n##\n\n\n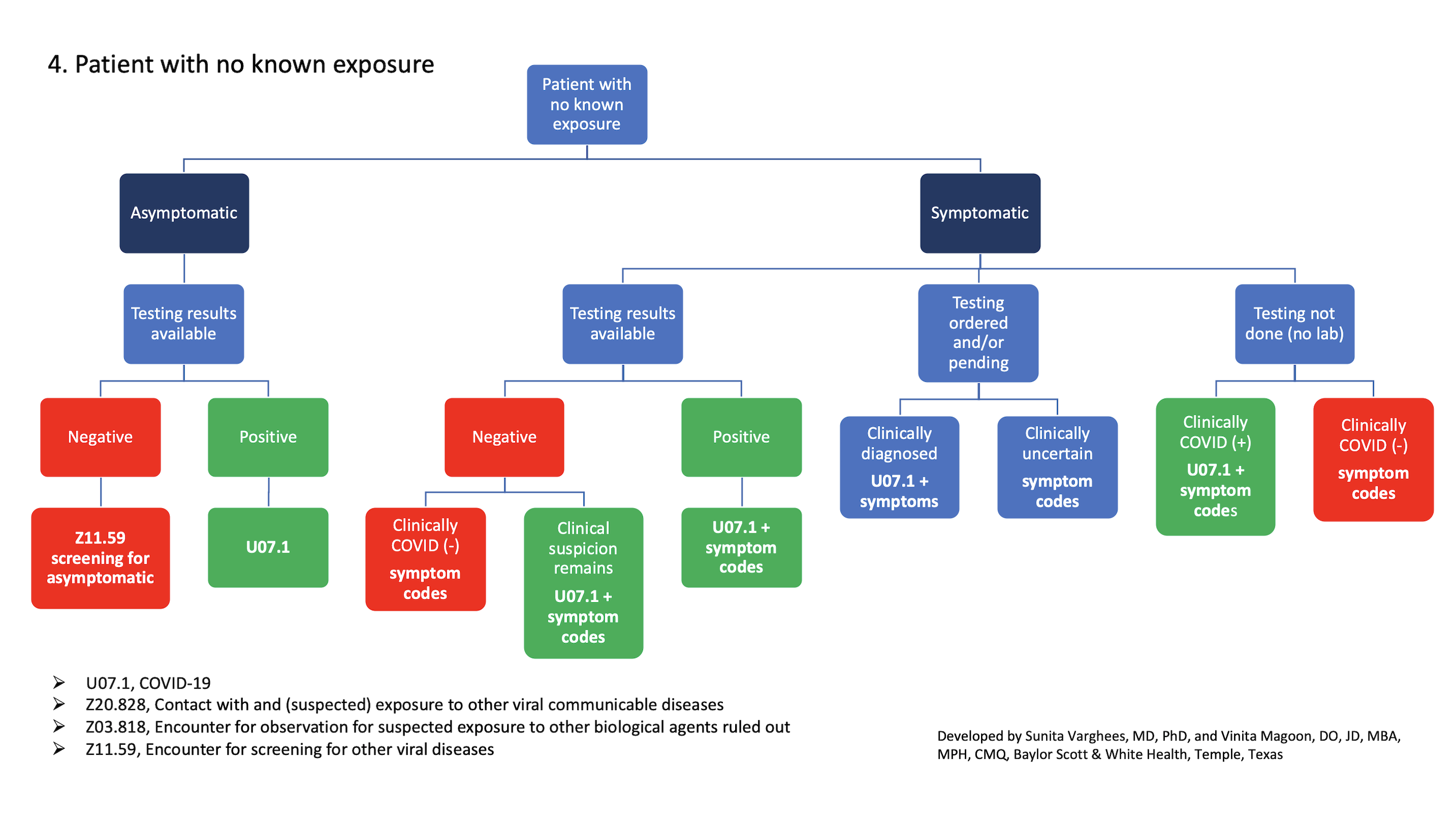\n\n\n\n## Mobility data\n\n\n::: {.cell layout-align=\"center\"}\n\n:::\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code}\nbigtree <- tree(Mobility ~ ., data = mob)\nsmalltree <- prune.tree(bigtree, k = .09)\ndraw.tree(smalltree, digits = 2)\n```\n\n::: {.cell-output-display}\n{fig-align='center'}\n:::\n:::\n\n\nThis is called the [dendrogram]{.secondary}\n\n\n## Partition view\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code}\nmob$preds <- predict(smalltree)\npar(mfrow = c(1, 2), mar = c(5, 3, 0, 0))\ndraw.tree(smalltree, digits = 2)\ncols <- viridisLite::viridis(20, direction = -1)[cut(log(mob$Mobility), 20)]\nplot(mob$Black, mob$Commute,\n pch = 19, cex = .4, bty = \"n\", las = 1, col = cols,\n ylab = \"Commute time\", xlab = \"% Black\"\n)\npartition.tree(smalltree, add = TRUE, ordvars = c(\"Black\", \"Commute\"))\n```\n\n::: {.cell-output-display}\n{fig-align='center'}\n:::\n:::\n\n\n\nWe predict all observations in a region with the same value. \n$\\bullet$ The three regions correspond to the leaves of the tree.\n\n\n##\n\n\n::: {.cell layout-align=\"center\"}\n\n```{.r .cell-code}\ndraw.tree(bigtree, digits = 2)\n```\n\n::: {.cell-output-display}\n{fig-align='center'}\n:::\n:::\n\n\n\n[Terminology]{.secondary}\n\nWe call each split or end point a node. Each terminal node is referred to as a leaf. \n\nThe interior nodes lead to branches. \n\n\n## Advantages and disadvantages of trees\n\n🎉 Trees are very easy to explain (much easier than even linear regression). \n\n🎉 Some people believe that decision trees mirror human decision. \n\n🎉 Trees can easily be displayed graphically no matter the dimension of the data.\n\n🎉 Trees can easily handle qualitative predictors without the need to create dummy variables.\n\n💩 Trees aren't very good at prediction.\n\n💩 Full trees badly overfit, so we \"prune\" them using CV \n\n. . .\n\n[We'll talk more about trees next module for Classification.]{.hand}\n\n# Next time ... {background-image=\"gfx/proforhobo.png\" background-opacity=.3}\n\n\nModule 3\n\nClassification\n\n", | ||
| "supporting": [ | ||
| "13-gams-trees_files" | ||
| ], | ||
| "filters": [ | ||
| "rmarkdown/pagebreak.lua" | ||
| ], | ||
| "includes": { | ||
| "include-after-body": [ | ||
| "\n<script>\n // htmlwidgets need to know to resize themselves when slides are shown/hidden.\n // Fire the \"slideenter\" event (handled by htmlwidgets.js) when the current\n // slide changes (different for each slide format).\n (function () {\n // dispatch for htmlwidgets\n function fireSlideEnter() {\n const event = window.document.createEvent(\"Event\");\n event.initEvent(\"slideenter\", true, true);\n window.document.dispatchEvent(event);\n }\n\n function fireSlideChanged(previousSlide, currentSlide) {\n fireSlideEnter();\n\n // dispatch for shiny\n if (window.jQuery) {\n if (previousSlide) {\n window.jQuery(previousSlide).trigger(\"hidden\");\n }\n if (currentSlide) {\n window.jQuery(currentSlide).trigger(\"shown\");\n }\n }\n }\n\n // hookup for slidy\n if (window.w3c_slidy) {\n window.w3c_slidy.add_observer(function (slide_num) {\n // slide_num starts at position 1\n fireSlideChanged(null, w3c_slidy.slides[slide_num - 1]);\n });\n }\n\n })();\n</script>\n\n" | ||
| ] | ||
| }, | ||
| "engineDependencies": {}, | ||
| "preserve": {}, | ||
| "postProcess": true | ||
| } | ||
| } |
Oops, something went wrong.