-
Notifications
You must be signed in to change notification settings - Fork 10
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
2 changed files
with
17 additions
and
44 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -2,55 +2,42 @@ | |
|
|
||
| <u>D</u>ata-Driven <u>O</u>ptimization of <u>M</u>ass <u>S</u>pectrometry Methods | ||
|
|
||
|  | ||
|  | ||
|  | ||
|
|
||
| * [Website](https://do-ms.slavovlab.net) | ||
| * [Get started now](#getting-started) | ||
| * [Download](https://github.com/SlavovLab/DO-MS/releases/latest) | ||
| * [Manuscript](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039) | ||
| * [BioArxiv Preprint](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1) | ||
|
|
||
| <img src="https://do-ms.slavovlab.net/assets/images/TOC_Graphic_noOutline_abstract_v18.png" width="70%"> | ||
| <img src="https://do-ms.slavovlab.net/assets/images/do-ms-dia_title_v2.png" width="70%"> | ||
|
|
||
|
|
||
| ## Getting Started | ||
|
|
||
| Please read our detailed getting started guides: | ||
| * [Getting started on the application](https://do-ms.slavovlab.net/docs/getting-started-application) | ||
| * [Getting started on the command-line](https://do-ms.slavovlab.net/docs/getting-started-command-line) | ||
| * [Getting started with DIA preprocessing](https://do-ms.slavovlab.net/docs/getting-started-preprocessing) | ||
| * [Getting started with DIA reports](https://do-ms.slavovlab.net/docs/getting-started-dia-app) | ||
| * [Getting started with DDA reports](https://do-ms.slavovlab.net/docs/getting-started-dda-app) | ||
|
|
||
| ### Requirements | ||
|
|
||
| This application has been tested on R >= 4.0.0 and R <= 4.0.2, OSX 10.14 / Windows 7/8/10. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and MaxQuant < 2.0 | ||
| This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10/11. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and DIA-NN > 1.8.1. | ||
|
|
||
| The application suffers from visual glitches when displayed on unsupported older browsers (such as IE9 commonly packaged with RStudio on Windows). Please use IE >= 11, Firefox, or Chrome for the best user experience. | ||
|
|
||
| ### Installation | ||
|
|
||
| Install this application by downloading it from the [release page](https://github.com/SlavovLab/DO-MS/releases/latest). | ||
|
|
||
| ### Running | ||
| ### Running the Interactive Application | ||
|
|
||
| The easiest way to run the app is directly through RStudio, by opening the `DO-MS.Rproj` Rproject file | ||
|
|
||
| <img src="https://github.com/SlavovLab/DO-MS/raw/master/docs/assets/images/do-ms-proj.png" height="100px"> | ||
| 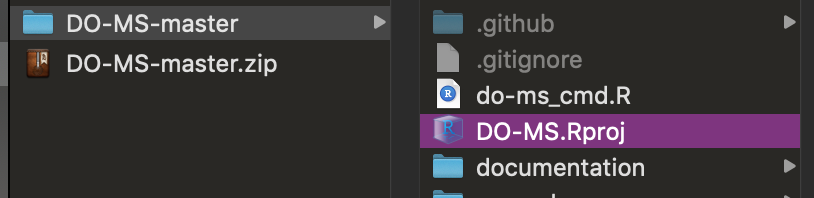{: width="70%" .center-image} | ||
|
|
||
| and clicking the "Run App" button at the top of the application, after opening the `server.R` file. We recommend checking the "Run External" option to open the application in your default browser instead of the RStudio Viewer. | ||
|
|
||
| <img src="https://github.com/SlavovLab/DO-MS/raw/master/docs/assets/images/do-ms-run.png" height="100px"> | ||
| 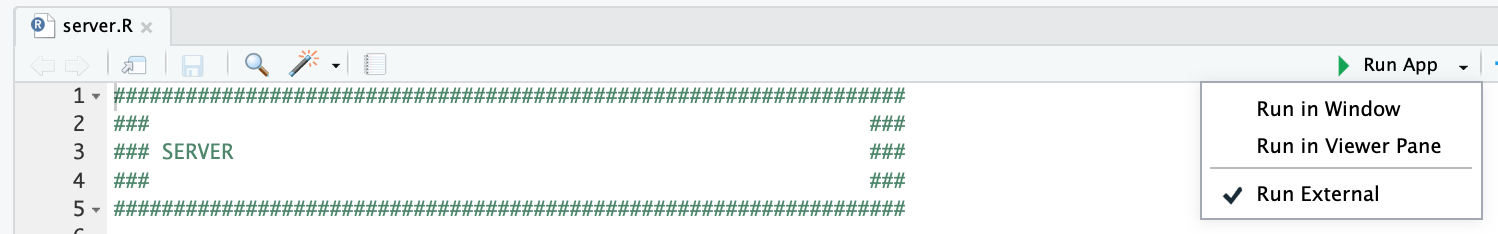{: width="70%" .center-image} | ||
|
|
||
| You can also start the application by running the `start_server.R` script. | ||
|
|
||
| ### Automated Report Generation | ||
|
|
||
| You can automatically generate PDF/HTML reports without having to launch the server by running the `do-ms_cmd.R` script, like so: | ||
|
|
||
| ``` | ||
| $ Rscript do-ms_cmd.R config_file.yaml | ||
| ``` | ||
|
|
||
| This requires a configuration file, and you can [find an example one here](https://github.com/SlavovLab/DO-MS/blob/master/example/config_file.yaml). See [Automating Report Generation](https://do-ms.slavovlab.net/docs/automation) for more details and instructions. | ||
|
|
||
| ### Customization | ||
|
|
||
| DO-MS is designed to be easily user-customizable for in-house proteomics workflows. Please see [Building Your Own Modules](https://do-ms.slavovlab.net/docs/build-your-own) for more details. | ||
|
|
@@ -59,9 +46,9 @@ DO-MS is designed to be easily user-customizable for in-house proteomics workflo | |
|
|
||
| Please see [Hosting as a Server](https://do-ms.slavovlab.net/docs/hosting-as-server) for more details. | ||
|
|
||
| ### Search Engines Other Than MaxQuant | ||
| ### Supporting other Search Engines | ||
|
|
||
| This application is currently maintained for MaxQuant >= 1.6.0.16. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details. | ||
| This application is currently maintained for (MaxQuant)[https://www.nature.com/articles/nbt.1511] >= 1.6.0.16 and (DIA-NN)[https://www.nature.com/articles/s41592-019-0638-x] >= 1.8. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details. | ||
|
|
||
| ### Can I use this for Metabolomics, Lipidomics, etc... ? | ||
|
|
||
|
|
@@ -71,13 +58,8 @@ While the base library of modules are based around bottom-up proteomics by LC-MS | |
|
|
||
| ## About the project | ||
|
|
||
| <!-- | ||
| DO-MS is a project... | ||
| --> | ||
|
|
||
| The manuscript for this tool is published at the Journal of Proteome Research: [https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039) | ||
|
|
||
| The manuscript is also freely available on bioRxiv: [https://www.biorxiv.org/content/10.1101/512152v1](https://www.biorxiv.org/content/10.1101/512152v1). | ||
| The manuscript for the extended version 2.0 can be found on bioArxiv: [https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1) | ||
|
|
||
| Contact the authors by email: [nslavov\{at\}northeastern.edu](mailto:[email protected]). | ||
|
|
||
|
|
@@ -87,21 +69,11 @@ DO-MS is distributed by an [MIT license](https://github.com/SlavovLab/DO-MS/blob | |
|
|
||
| ### Contributing | ||
|
|
||
| Please feel free to contribute to this project by opening an issue or pull request. | ||
|
|
||
| <!-- | ||
| ### Data | ||
| All data used for the manuscript is available on [UCSD's MassIVE Repository](https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=ed5a1ab37dc34985bbedbf3d9a945535) | ||
| --> | ||
|
|
||
| <!-- | ||
| ### Figures/Analysis | ||
| Scripts for the figures in the DART-ID manuscript are available in a separate GitHub repository, [https://github.com/SlavovLab/DART-ID_2018](https://github.com/SlavovLab/DART-ID_2018) | ||
| --> | ||
| Please feel free to contribute to this project by opening an issue or pull request in the [GitHub repository](https://github.com/SlavovLab/DO-MS). | ||
|
|
||
| ------------- | ||
|
|
||
| ## Help! | ||
|
|
||
| For any bugs, questions, or feature requests, | ||
| For any bugs, questions, or feature requests, | ||
| please use the [GitHub issue system](https://github.com/SlavovLab/DO-MS/issues) to contact the developers. | ||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters