pynucastro is a python library for interactively creating and exploring nuclear reaction networks. It provides interfaces to nuclear reaction rate databases, including the JINA Reaclib nuclear reactions database.
The main features are:
-
Ability to create a reaction network based on a collection of rates, a set of nuclei, or an arbitrary filter applied to a library.
-
Interactive exploration of rates and networks in Jupyter notebooks.
-
Many different ways of visualizing a network.
-
An NSE solver to find the equilibrium abundance of a set of nuclei given a thermodynamic state.
-
Ability to write out python or C++ code needed to integrate the network.
-
Support for tabular weak rates.
-
Rate approximations and the derivation of reverse rates via detailed balance.
-
Easy access to nuclear properties, including T-dependent partition functions, spins, masses, etc.
Documentation for pynucastro is available here:
http://pynucastro.github.io/pynucastro/
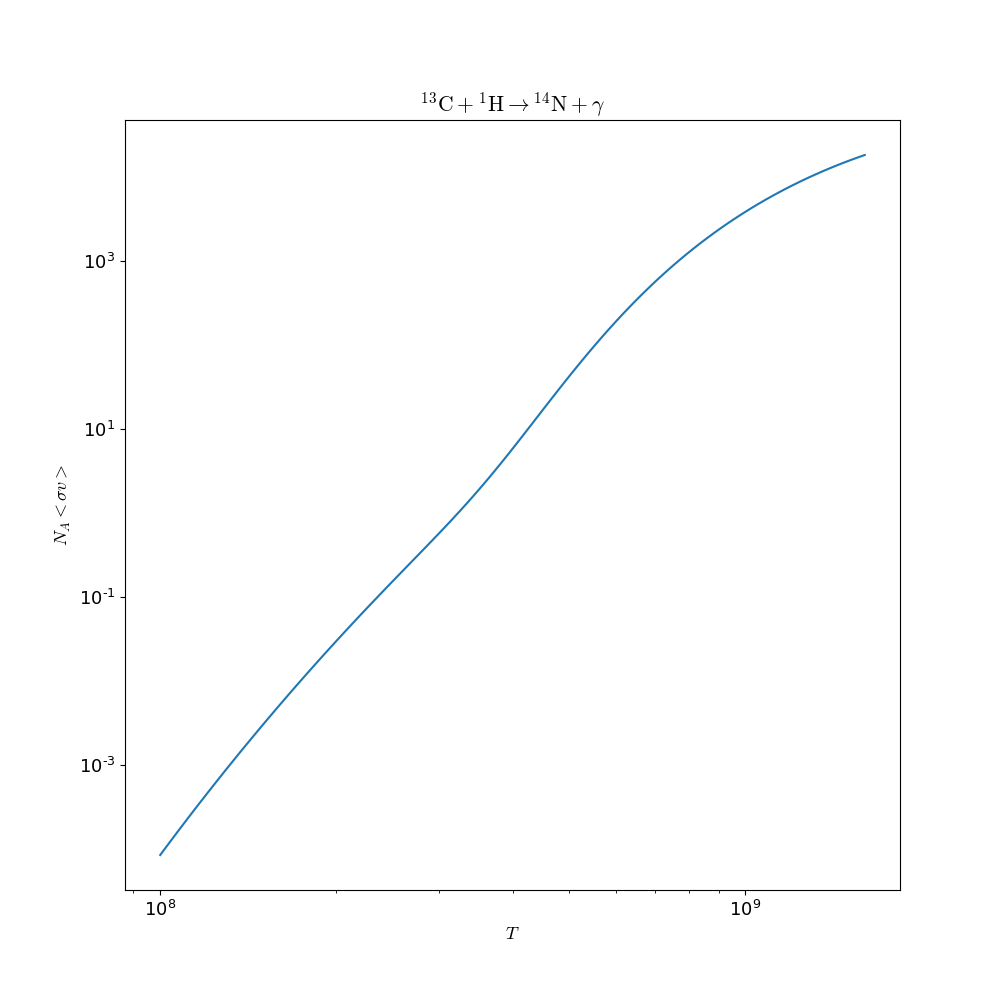
The following example reads in the ReacLib rate database and gets the rate for C13(p,g)N14 and evaluates it at a temperature of 1.e9 K and makes a plot of the T dependence:
In [1]: import pynucastro as pyna
In [2]: rl = pyna.ReacLibLibrary()
In [3]: c13pg = rl.get_rate_by_name("c13(p,g)n14")
In [4]: c13pg.eval(1.e9)
Out[4]: 3883.4778216250666
In [5]: fig = c13pg.plot()
In [6]: fig.savefig("c13pg.png")
An extensive demonstration of the capabilities of pynucastro is shown in this notebook:
We can also interactively explore a reaction network. Here's an example of hot-CNO with breakout reactions:
by increasing the temperature, you can see the transition from CNO to hot-CNO (proton captures on C13 become faster than the beta decay) and then the breakout of hot-CNO when the alpha capture on O14 becomes faster than its decay.
pynucastro is available on PyPI and can be installed via:
pip install pynucastro
Alternately, to install via source, you can do:
python setup.py install
for a systemwide install, or
python setup.py install --user
for a single-user install. This will put the pynucastro modules and library in the default location python searches for packages.
This package requires Python 3 (release 3.6 or later) and the following Python packages:
-
numpy -
sympy -
scipy -
matplotlib -
networkx -
ipywidgets -
numba -
setuptools_scm
To build the documentation or run the unit tests, sphinx and
pytest are additionally required along with some supporting
packages. See the included requirements.txt file for a list of these
packages and versions. To install the packages from the requirements
file, do:
pip install -r requirements.txt
Is important to stress out that all the dependencies should be
installed before pynucastro, otherwise the installation should be
repeated.
We use py.test to do unit tests. In pynucastro/, do:
pytest -v .
to see coverage, do:
pytest --cov=pynucastro .
to test the notebooks, do:
py.test --nbval examples
People who make a number of substantive contributions (new features, bug fixes, etc.) will be named "core developers" of pynucastro.
Core developers will be recognized in the following ways:
-
invited to the group's slack team
-
listed in the User's Guide and website as a core developer
-
listed in the author list on the Zenodo DOI for the project (as given in the .zenodo.json file)
-
invited to co-author general code papers / proceedings describing pynucastro. (Note: science papers that use pynucastro will always be left to the science paper lead author to determine authorship).
If a core developer is inactive for 3 years, we may reassess their status as a core developer.