Trying to reproduce BioBERT(paper link) results for the NER task. The original work of the paper is in Tensorflow. This repo is made using 🤗HuggingFace transformers library and Pytorch.
- Code and project workflow heavily inspired by Abhishek Thakur's tutorials.
- 🤗HuggingFace's Transformers documentation
- Evaluation details and other specifics taken from original paper
Run setup.sh to download the NER datasets of the paper and do preprocessing.
bash setup.shRun train.py to finetune the model on disease-datasets of the paper. Additionally to finetune on other datasets, make changes to config.py. There is an optional argument of secondary folderpath which can be when working on a remote server. When invoked, apart from saving the metadata and model in the local repository, it is also saved at a secondary path (For eg a G-drive path when working on a Google Colab server).
cd src
python train.py [-f(optional) "SECONDARY_FOLDERPATH"]The best model (according to val score) will be saved in models/ and metadata (containing label encoder object) will be saved in src/. The same will be done for the SECONDARY_FOLDERPATH if provided.
Make sure your trained model is in models/ and your metadata dict containing the LabelEncoder object is in src/. Run evaluate.py. There is optional argument -g which if invoked, will evaluate all metrics on exact entity-level matching; use python evaluate.py --help for more info on the arguments. You should see a classification report generated with the help of seqeval.
cd src
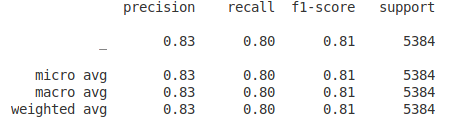
python evaluate.py [-g]Classification report of the NER task on a model finetuned for 10 epochs on the disease-datasets:
Here evaluation is done for single entity-type (disease) and hence the averages are the same.
- In the original paper, they have pretrained on biomedical datasets and then finetuned for downstream tasks. Here we only finetune a
bert-base-uncasedmodel without any pretraining. The model can be changed inconfig.pywith minimal changes tomodel.py. evaluate.pycontains custom methods by which we carry out entity-level NER exact evaluation (as stated in the paper). Meaning we first convert the predictions on Wordpiece tokens to word-level predictions, and then carry out exact matching to get NER metrics namely F1 score, precision and recall.