-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Added JSON formatting to RAG prompt builder
- Loading branch information
1 parent
430ca61
commit caff400
Showing
5 changed files
with
37 additions
and
8 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -165,3 +165,4 @@ metrics.txt | |
| metrics.png | ||
| gdrive-oauth.txt | ||
| /eval | ||
| .tmp/ | ||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
|
|
@@ -16,3 +16,4 @@ | |
| /eidc_rag_testset.csv | ||
| /eidc_rag_test_set.csv | ||
| /rag-pipeline.yml | ||
| /pipeline.yml | ||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
caff400There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
context_precision: 0.45696799875829536
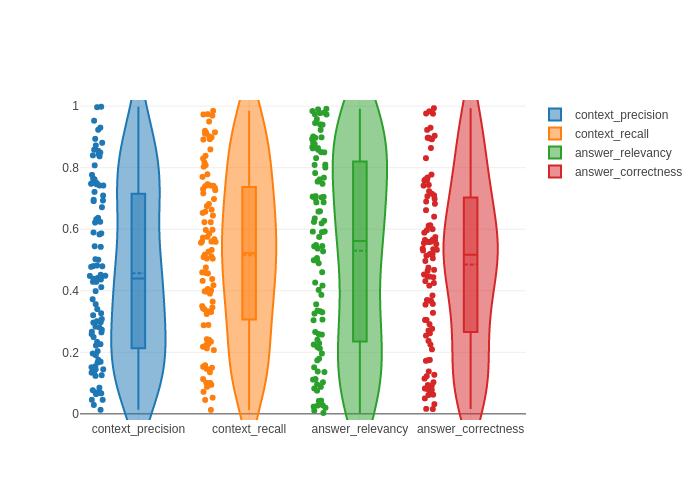
context_recall: 0.5150504867529684
answer_relevancy: 0.5303345339043942
answer_correctness: 0.4851447082020083