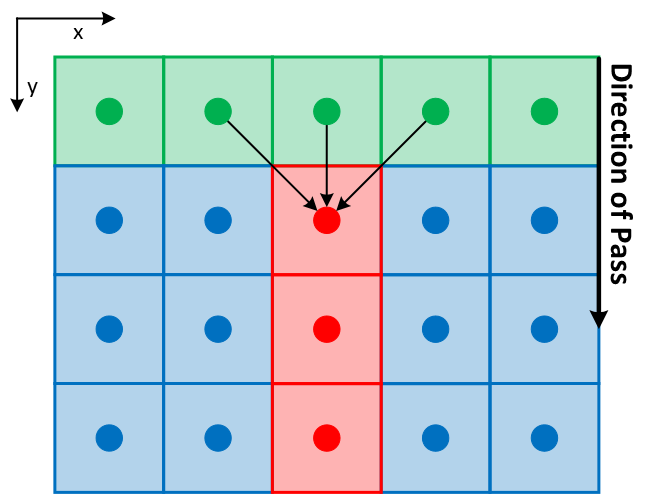
This repository provides CPU (OpenMP) and GPU (CUDA) implementations of Generalised Geodesic Distance Transform in PyTorch for 2D and 3D input data based on parallelisable raster scan ideas from [1]. It includes methods for computing Geodesic, Euclidean distance transform and mixture of both.
| 2D images, 1 of 4 passes | 3D volumes, 1 of 6 passes |
|---|---|
 |
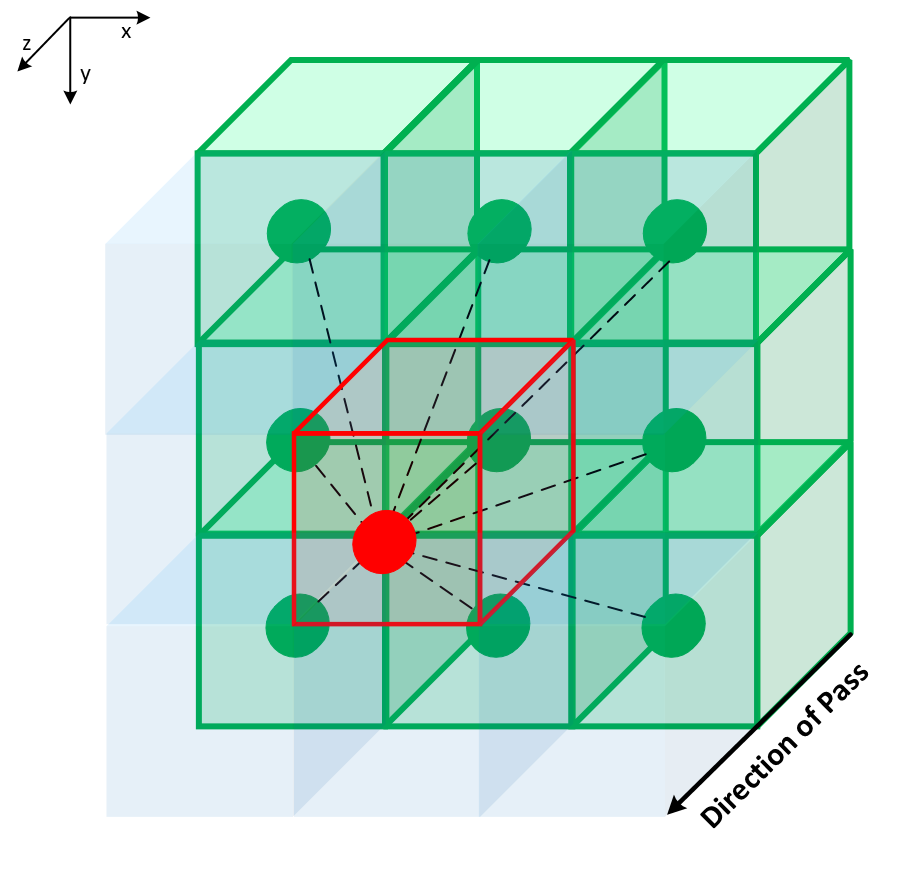 |
The above raster scan method can be parallelised for each row/plane on an available device (CPU or GPU). This leads to significant speed up as compared to existing non-parallelised raster scan implementations (e.g. https://github.com/taigw/GeodisTK). Python interface is provided (using PyTorch) for enabling its use in deep learning and image processing pipelines.
In addition, implementation of generalised version of Geodesic distance transforms along with Geodesic Symmetric Filtering (GSF) is provided for use in interactive segmentation methods, that were originally proposed in [1, 2, 5].
The raster scan based implementation provides a balance towards speed rather than accuracy of Geodesic distance transform and hence results in efficient hardware utilisation. On the other hand, in case of Euclidean distance transform, exact results can be achieved with other packages (albeit not on necessarilly on GPU) [6, 7, 8]
If you use this code in your research, then please consider citing:
Asad, Muhammad, Reuben Dorent, and Tom Vercauteren. "FastGeodis: Fast Generalised Geodesic Distance Transform." Journal of Open Source Software (JOSS), 2022. (paper link)
Bibtex:
@article{asad2022fastgeodis,
doi = {10.21105/joss.04532},
url = {https://doi.org/10.21105/joss.04532},
year = {2022},
publisher = {The Open Journal},
volume = {7},
number = {79},
pages = {4532},
author = {Muhammad Asad and Reuben Dorent and Tom Vercauteren},
title = {FastGeodis: Fast Generalised Geodesic Distance Transform},
journal = {Journal of Open Source Software}
}
The provided package can be installed using:
pip install FastGeodis
or
pip install git+https://github.com/masadcv/FastGeodis
or (on conda environments with existing installation of PyTorch with CUDA)
pip install FastGeodis --no-build-isolation
| Method | Description | Documentation |
|---|---|---|
| Fast Generalised Geodesic Distance 2D | Paralellised generalised geodesic distance transform for CPU/GPU [1] | FastGeodis.generalised_geodesic2d |
| Fast Generalised Geodesic Distance 3D | Paralellised generalised geodesic distance transform for CPU/GPU [1] | FastGeodis.generalised_geodesic3d |
| Fast Signed Generalised Geodesic Distance 2D | Paralellised signed generalised geodesic distance transform for CPU/GPU [1] | FastGeodis.signed_generalised_geodesic2d |
| Fast Signed Generalised Geodesic Distance 3D | Paralellised signed generalised geodesic distance transform for CPU/GPU [1] | FastGeodis.signed_generalised_geodesic3d |
| Fast Geodesic Symmetric Filtering 2D | Paralellised geodesic symmetric filtering for CPU/GPU [2] | FastGeodis.GSF2d |
| Fast Geodesic Symmetric Filtering 3D | Paralellised geodesic symmetric filtering for CPU/GPU [2] | FastGeodis.GSF3d |
| Method | Description | Documentation |
|---|---|---|
| Toivanen's Generalised Geodesic Distance 2D | Toivanen's generalised geodesic distance transform for CPU [9] | FastGeodis.generalised_geodesic2d_toivanen |
| Toivanen's Generalised Geodesic Distance 3D | Toivanen's generalised geodesic distance transform for CPU [9] | FastGeodis.generalised_geodesic3d_toivanen |
| Toivanen's Signed Generalised Geodesic Distance 2D | Toivanen's signed generalised geodesic distance transform for CPU [9] | FastGeodis.signed_generalised_geodesic2d_toivanen |
| Toivanen's Signed Generalised Geodesic Distance 3D | Toivanen's signed generalised geodesic distance transform for CPU [9] | FastGeodis.signed_generalised_geodesic3d_toivanen |
| Toivanen's Geodesic Symmetric Filtering 2D | Toivanen's geodesic symmetric filtering for CPU [2, 9] | FastGeodis.GSF2d_toivanen |
| Toivanen's Geodesic Symmetric Filtering 3D | Toivanen's geodesic symmetric filtering for CPU [2, 9] | FastGeodis.GSF3d_toivanen |
| Method | Description | Documentation |
|---|---|---|
| Pixel Queue Geodesic Distance 2D | Pixel Queue geodesic distance transform for CPU [11] | FastGeodis.geodesic2d_fastmarch |
| Pixel Queue Geodesic Distance 3D | Pixel Queue geodesic distance transform for CPU [11] | FastGeodis.geodesic3d_pixelqueue |
| Pixel Queue Signed Geodesic Distance 2D | Pixel Queue signed geodesic distance transform for CPU [11] | FastGeodis.signed_geodesic2d_pixelqueue |
| Pixel Queue Signed Geodesic Distance 3D | Pixel Queue signed geodesic distance transform for CPU [11] | FastGeodis.signed_geodesic3d_pixelqueue |
| Pixel Queue Geodesic Symmetric Filtering 2D | Pixel Queue geodesic symmetric filtering for CPU [2, 11] | FastGeodis.GSF2d_pixelqueue |
| Pixel Queue Geodesic Symmetric Filtering 3D | Pixel Queue geodesic symmetric filtering for CPU [2, 11] | FastGeodis.GSF3d_pixelqueue |
| Method | Description | Documentation |
|---|---|---|
| Fast Marching Geodesic Distance 2D | Fast Marching geodesic distance transform for CPU [9] | FastGeodis.geodesic2d_fastmarch |
| Fast Marching Geodesic Distance 3D | Fast Marching geodesic distance transform for CPU [9] | FastGeodis.geodesic3d_fastmarch |
| Fast Marching Signed Geodesic Distance 2D | Fast Marching signed geodesic distance transform for CPU [9] | FastGeodis.signed_geodesic2d_fastmarch |
| Fast Marching Signed Geodesic Distance 3D | Fast Marching signed geodesic distance transform for CPU [9] | FastGeodis.signed_geodesic3d_fastmarch |
| Fast Marching Geodesic Symmetric Filtering 2D | Fast Marching geodesic symmetric filtering for CPU [2, 9] | FastGeodis.GSF2d_fastmarch |
| Fast Marching Geodesic Symmetric Filtering 3D | Fast Marching geodesic symmetric filtering for CPU [2, 9] | FastGeodis.GSF3d_fastmarch |
The following demonstrates a simple example showing FastGeodis usage:
To compute Geodesic Distance Transform:
device = "cuda" if torch.cuda.is_available() else "cpu"
image = np.asarray(Image.open("data/img2d.png"), np.float32)
image_pt = torch.from_numpy(image).unsqueeze_(0).unsqueeze_(0)
image_pt = image_pt.to(device)
mask_pt = torch.ones_like(image_pt)
mask_pt[..., 100, 100] = 0
v = 1e10
# lamb = 0.0 (Euclidean) or 1.0 (Geodesic) or (0.0, 1.0) (mixture)
lamb = 1.0
iterations = 2
geodesic_dist = FastGeodis.generalised_geodesic2d(
image_pt, mask_pt, v, lamb, iterations
)
geodesic_dist = np.squeeze(geodesic_dist.cpu().numpy())To compute Euclidean Distance Transform:
device = "cuda" if torch.cuda.is_available() else "cpu"
image = np.asarray(Image.open("data/img2d.png"), np.float32)
image_pt = torch.from_numpy(image).unsqueeze_(0).unsqueeze_(0)
image_pt = image_pt.to(device)
mask_pt = torch.ones_like(image_pt)
mask_pt[..., 100, 100] = 0
v = 1e10
# lamb = 0.0 (Euclidean) or 1.0 (Geodesic) or (0.0, 1.0) (mixture)
lamb = 0.0
iterations = 2
euclidean_dist = FastGeodis.generalised_geodesic2d(
image_pt, mask_pt, v, lamb, iterations
)
euclidean_dist = np.squeeze(euclidean_dist.cpu().numpy())For more usage examples see:
| Description | Python | Colab link |
|---|---|---|
| Simple 2D Geodesic and Euclidean Distance | samples/simpledemo2d.py |
|
| Simple Signed 2D Geodesic and Euclidean Distance | samples/simpledemo2d_signed.py |
|
| Simple 3D Geodesic and Euclidean Distance | samples/simpledemo3d.py |
|
| Simple Signed 3D Geodesic and Euclidean Distance | samples/simpledemo3d_signed.py |
|
| 2D Geodesic Distance | samples/demo2d.py |
|
| 2D Signed Geodesic Distance | samples/demo2d_signed.py |
|
| 3D Geodesic Distance | samples/demo3d.py |
|
| 3D Signed Geodesic Distance | samples/demo3d_signed.py |
|
| 2D GSF Segmentation Smoothing | samples/demoGSF2d_SmoothingSegExample.ipynb |
A number of unittests are provided, which can be run as:
pip install -r requirements-dev.txt
python -m unittest
Further details of each function implemented in FastGeodis can be accessed at the documentation hosted at: https://fastgeodis.readthedocs.io.
Spotted a bug or have a feature request to improve the package? We would love to have your input! See our guidelines for contributing.
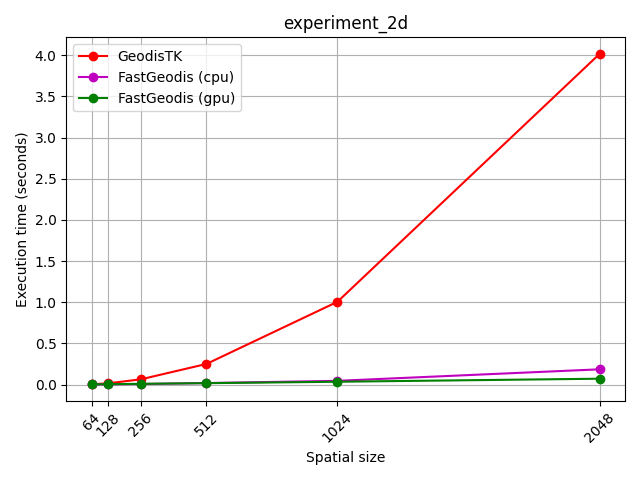
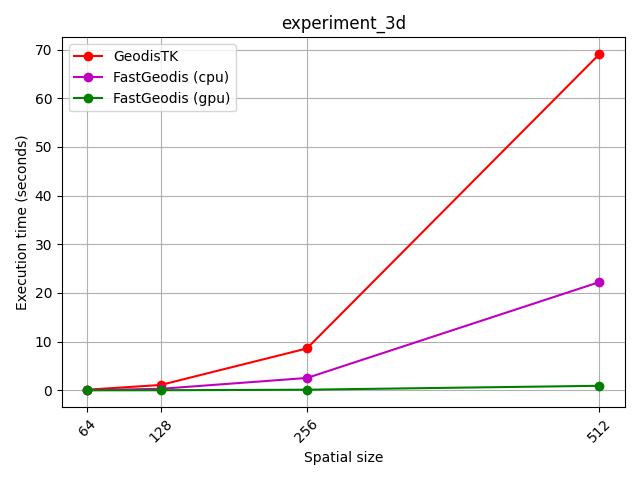
FastGeodis (CPU/GPU) is compared with existing GeodisTK (https://github.com/taigw/GeodisTK) in terms of execution speed as well as accuracy.
| 2D images | 3D volumes |
|---|---|
 |
 |
2D case
| Qualitative Comparison | Quantitative (joint histogram) |
|---|---|
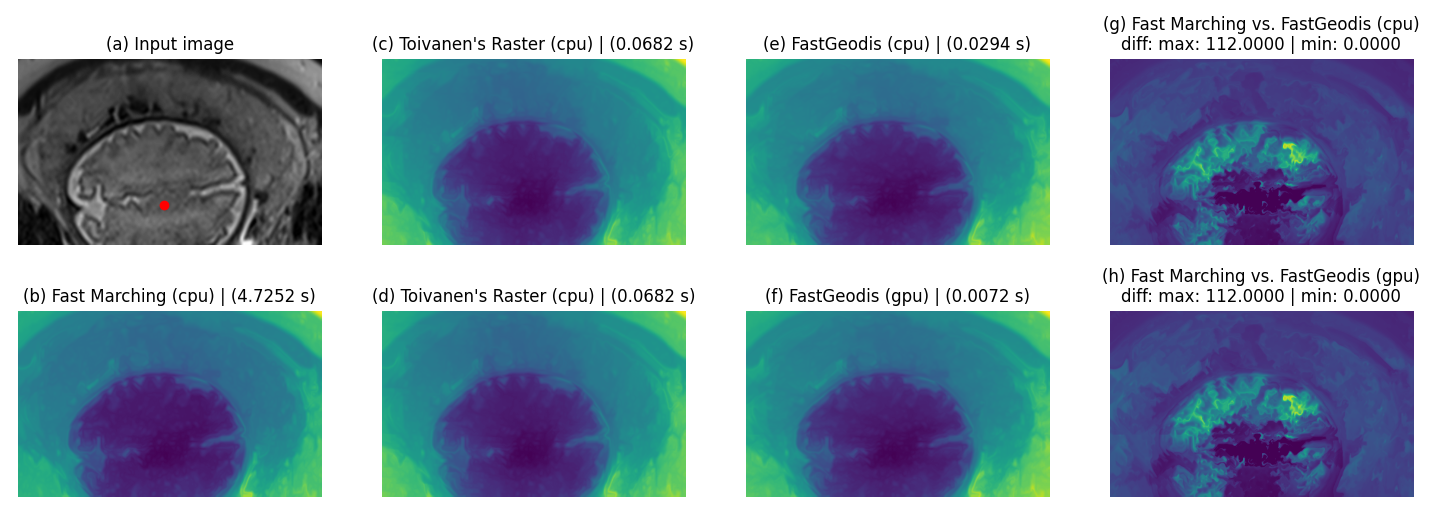 |
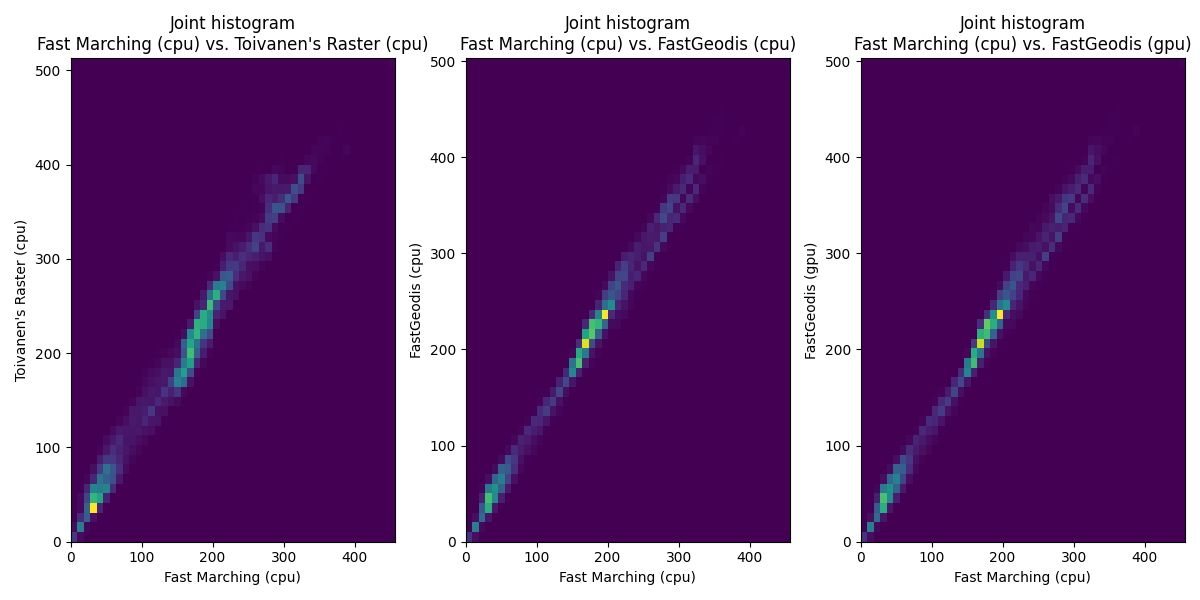 |
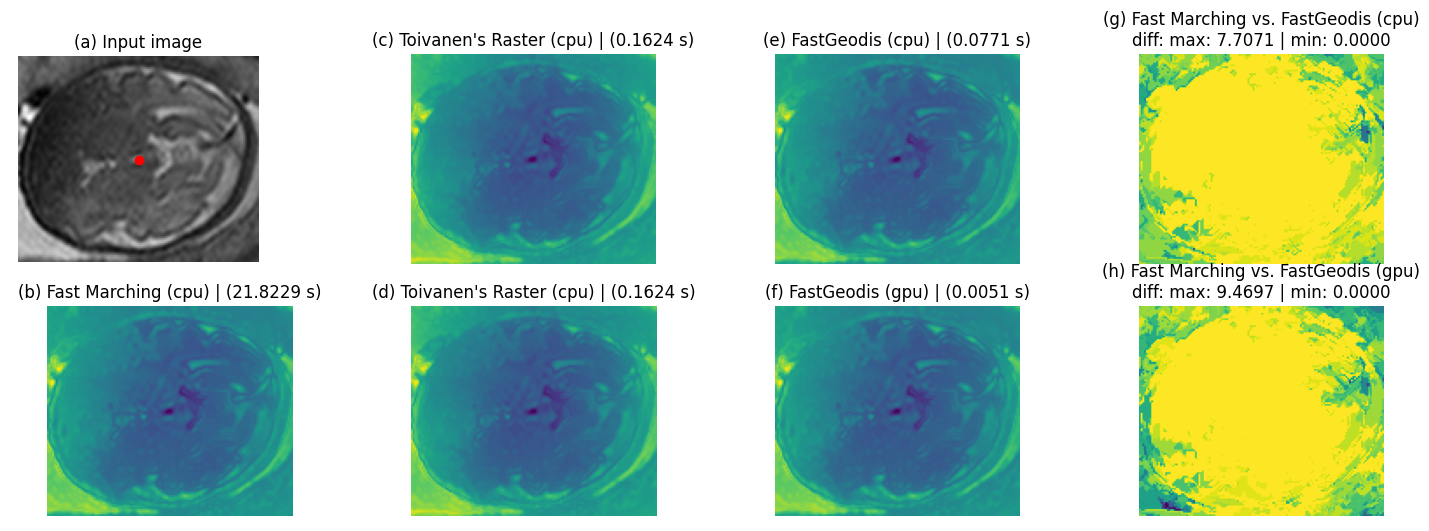
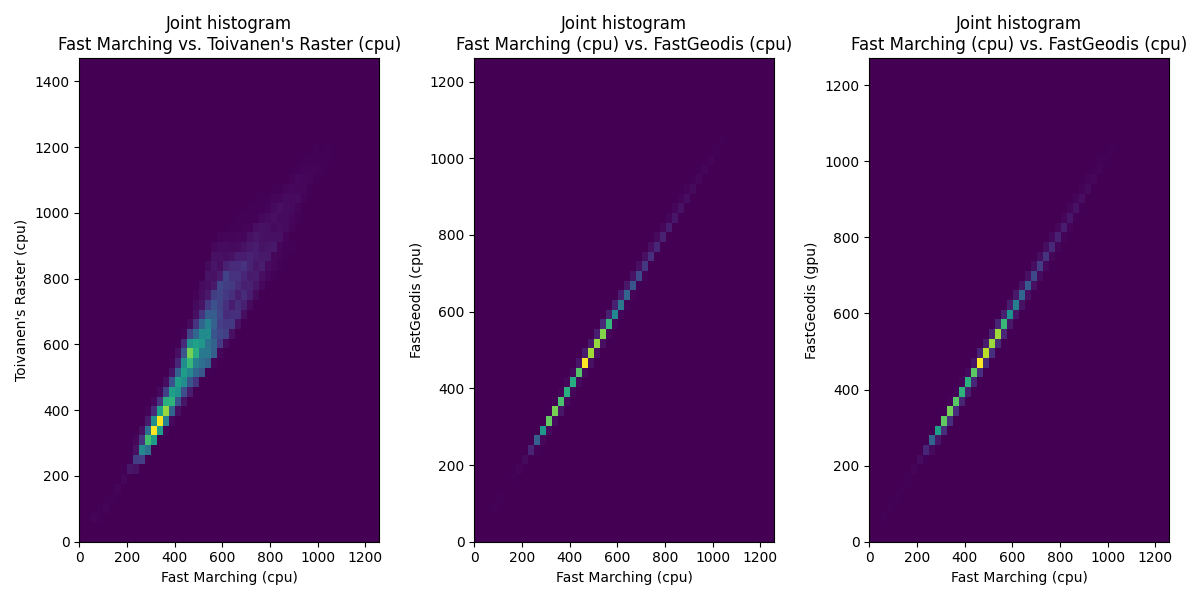
3D case
| Qualitative Comparison | Quantitative (joint histogram) |
|---|---|
 |
 |
-
[1] Criminisi, Antonio, Toby Sharp, and Khan Siddiqui. "Interactive Geodesic Segmentation of n-Dimensional Medical Images on the Graphics Processor." Radiological Society of North America (RSNA), 2009. [pdf, ppt, url]
-
[2] Criminisi, Antonio, Toby Sharp, and Andrew Blake. "Geos: Geodesic image segmentation." European Conference on Computer Vision. Springer, Berlin, Heidelberg, 2008. [doi]
-
[3] Weber, Ofir, et al. "Parallel algorithms for approximation of distance maps on parametric surfaces." ACM Transactions on Graphics (TOG), (2008). [doi]
-
[4] GeodisTK: https://github.com/taigw/GeodisTK
-
[5] Criminisi, Antonio, Toby Sharp, Carsten Rother, and Patrick Pérez. "Geodesic image and video editing." ACM Trans. Graph. 29, no. 5 (2010): 134-1. [doi]
-
[6] https://github.com/seung-lab/euclidean-distance-transform-3d
-
[7] https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.distance_transform_edt.html
-
[8] https://www.tensorflow.org/addons/api_docs/python/tfa/image/euclidean_dist_transform
-
[9] Toivanen, Pekka J. "New geodosic distance transforms for gray-scale images." Pattern Recognition Letters 17.5 (1996): 437-450.
-
[10] Sethian, James A. "Fast marching methods." SIAM review 41.2 (1999): 199-235.
-
[11] Ikonen, L., & Toivanen, P. (2007). Distance and nearest neighbor transforms on gray-level surfaces. Pattern Recognition Letters, 28(5), 604-612. [doi]



