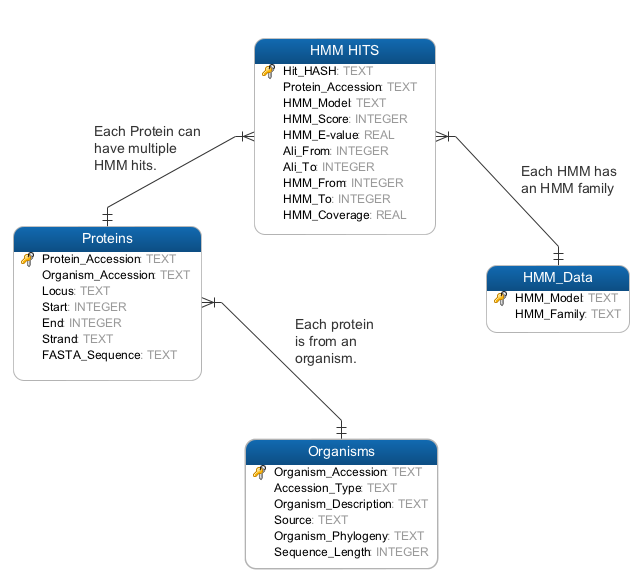
Software for searching, extracting and storing proteins that match specific Hidden Markov Models. Hits generated by HMMER's hmmsearch are filtered and stored in a SQLite database. After running the HMMER-DB suite the database will contain:
- Information about HMM hits�: Protein accession, E-Value, score coverage, etc.
- Information about hit proteins: Protein accession, Organism accession, locus, location, etc.
- Information about organisms with hits: Organism accession, description and phylogeny, etc.
Optionally we have also included a table called HMM_Data that lets you group certain HMMs into families.
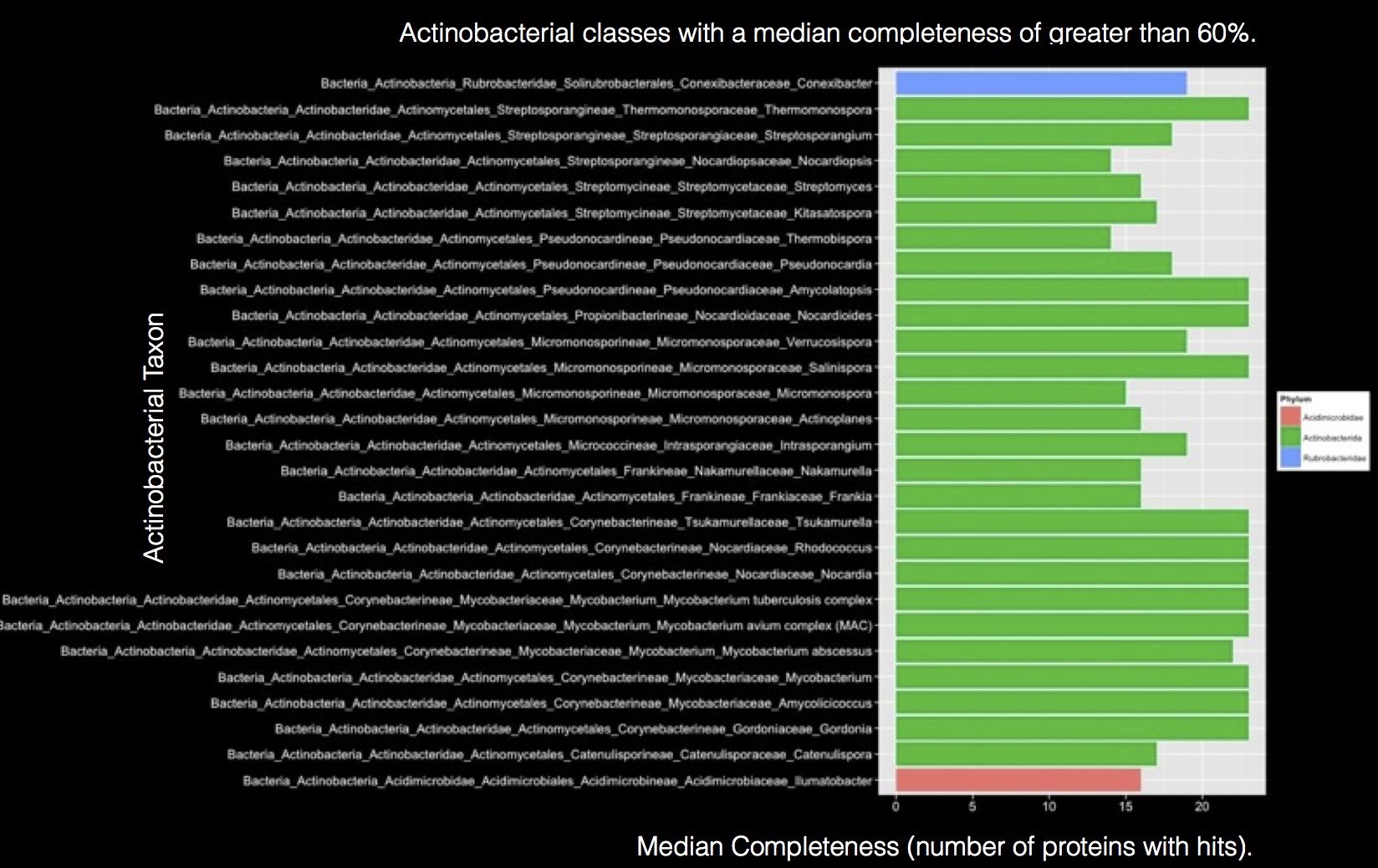
Once the the database is setup one can easily query it using SQL to gather information about a protein's or group of proteins' potential distribution across multiple taxon. We have also included a few R scripts to help with visualization. Here is an example:
To set up the database is a three part process.
- Run GenbankToFASTAandOrganismTable.py on a series of Genbank files containing the organisms you wish to search with your HMMs.
- Run SearchExtractLoad.py on the resulting FASTA files and csv table from GenbankToFASTAandOrganismTable.py. SearchExtractLoad.py will also run hmmsearch, extract hits and load all the hits and other the information into the database.
- Query to your heart's content.