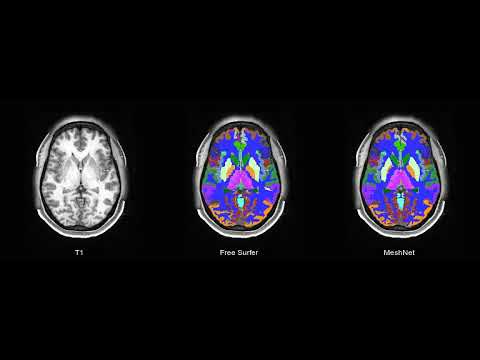
This repository contains a PyTorch implementation of MeshNet architecture. MeshNet is a volumetric convolutional neural network for image segmentation (focused on brain imaging application) based on dilated kernels [1].
This code provides a framework for training and evaluating a model for segmentation of a T1 (+ optional T2) into a 104 class brain atlas. It is a modification of our previous work [3].
- Prepare T1 or T2 input with mri_convert from FreeSurfer (https://surfer.nmr.mgh.harvard.edu/) conform T1 to 1mm voxel size in coronal slice direction with side length 256. You can skip this step if your T1 image is already with slice thickness 1mm x 1mm x 1mm and 256 x 256 x 256.
mri_convert [brainDir]/t1.nii [brainDir]/T1.nii.gz -c
- Prepare labels from aparc+aseg.nii.gz using:
python prepare_data.py --brains_list [brains_lits.txt]
To train the model use the following command:
python main.py --model ./models_configurations/MeshNet_104_38.yml --train_path ./folds/hcp_example/train.txt --validation_path ./folds/hcp_example/validation.txt
We also support Visdom (https://github.com/facebookresearch/visdom) monitoring during training. To use it use arguments:
--visdom --visdom_server [visdom_server_ip] --visdom_port [visdom_server_port]
To evaluate the model use the following command:
python evaluation.py --models_file [models_list.txt] --evaluation_path folds/hcp_example/test.txt
- Install PyTorch https://pytorch.org/get-started/locally/
- Install other dependencies
pip install -r requirements.txt
This work is supported by grants IIS-1318759, R01EB020407, R01EB006841, P20GM103472, P30GM122734.
Data were provided [in part] by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
[1] https://arxiv.org/abs/1511.07122 Multi-Scale Context Aggregation by Dilated Convolutions. Fisher Yu, Vladlen Koltun
[2] https://arxiv.org/abs/1612.00940 End-to-end learning of brain tissue segmentation from imperfect labeling. Alex Fedorov, Jeremy Johnson, Eswar Damaraju, Alexei Ozerin, Vince D. Calhoun, Sergey M. Plis
[3] https://arxiv.org/abs/1711.00457 Almost instant brain atlas segmentation for large-scale studies. Alex Fedorov, Eswar Damaraju, Vince Calhoun, Sergey Plis
[4] http://www.humanconnectomeproject.org/ Human Connectome Project
Brain Atlas segmentation with brainchop.org
To get brain atlas segmentation ([3]) you don't need to run any code. Just sign up at brainchop.org, upload your 3T MRI T1 image and get brain atlas in 1-2 minutes.
Watch video with example of brain atlas segmentation.
Torch Implementation for brain tissue segmentation https://github.com/Entodi/MeshNet
| T1 MRI | FreeSurfer | MeshNet |
|---|---|---|
 |
 |
 |
 |
 |
 |
 |
 |
 |
If you have any questions about implementation and training, don't hesitate to either open an issue here or send an email to [email protected].