-
Notifications
You must be signed in to change notification settings - Fork 2
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Signed-off-by: Zhiyuan Chen <[email protected]>
- Loading branch information
1 parent
580bb01
commit 84a494c
Showing
10 changed files
with
627 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,9 @@ | ||
| --- | ||
| authors: | ||
| - Zhiyuan Chen | ||
| date: 2024-05-04 | ||
| --- | ||
|
|
||
| # EternaBench-CM | ||
|
|
||
| --8<-- "multimolecule/datasets/eternabench_cm/README.md:21:" |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,9 @@ | ||
| --- | ||
| authors: | ||
| - Zhiyuan Chen | ||
| date: 2024-05-04 | ||
| --- | ||
|
|
||
| # EternaBench-External | ||
|
|
||
| --8<-- "multimolecule/datasets/eternabench_external/README.md:21:" |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,9 @@ | ||
| --- | ||
| authors: | ||
| - Zhiyuan Chen | ||
| date: 2024-05-04 | ||
| --- | ||
|
|
||
| # EternaBench-Switch | ||
|
|
||
| --8<-- "multimolecule/datasets/eternabench_switch/README.md:21:" |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,110 @@ | ||
| --- | ||
| language: rna | ||
| tags: | ||
| - Biology | ||
| - RNA | ||
| license: | ||
| - agpl-3.0 | ||
| size_categories: | ||
| - 1K<n<10K | ||
| task_categories: | ||
| - text-generation | ||
| - fill-mask | ||
| task_ids: | ||
| - language-modeling | ||
| - masked-language-modeling | ||
| pretty_name: EternaBench-CM | ||
| library_name: multimolecule | ||
| --- | ||
|
|
||
| # EternaBench-CM | ||
|
|
||
| 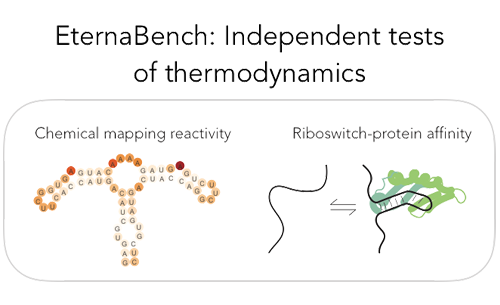 | ||
|
|
||
| EternaBench-CM is a synthetic RNA dataset comprising 12,711 RNA constructs that have been chemically mapped using SHAPE and MAP-seq methods. | ||
| These RNA sequences are probed to obtain experimental data on their nucleotide reactivity, which indicates whether specific regions of the RNA are flexible or structured. | ||
| The dataset provides high-resolution, large-scale data that can be used for studying RNA folding and stability. | ||
|
|
||
| ## Disclaimer | ||
|
|
||
| This is an UNOFFICIAL release of the [EternaBench-CM](https://github.com/eternagame/EternaBench) by Hannah K. Wayment-Steele, et al. | ||
|
|
||
| **The team releasing EternaBench-CM did not write this dataset card for this dataset so this dataset card has been written by the MultiMolecule team.** | ||
|
|
||
| ## Dataset Description | ||
|
|
||
| - **Homepage**: https://multimolecule.danling.org/datasets/eternabench_cm | ||
| - **datasets**: https://huggingface.co/datasets/multimolecule/eternabench-cm | ||
| - **Point of Contact**: [Rhiju Das](https://biochemistry.stanford.edu/people/rhiju-das/) | ||
|
|
||
| The dataset includes a large set of synthetic RNA sequences with experimental chemical mapping data, which provides a quantitative readout of RNA nucleotide reactivity. These data are ensemble-averaged and serve as a critical benchmark for evaluating secondary structure prediction algorithms in their ability to model RNA folding dynamics. | ||
|
|
||
| ## Example Entry | ||
|
|
||
| | index | design | sequence | secondary_structure | reactivity | errors | signal_to_noise | | ||
| | -------- | ---------------------- | ---------------- | ------------------- | -------------------------- | --------------------------- | --------------- | | ||
| | 769337-1 | d+m plots weaker again | GGAAAAAAAAAAA... | ................ | [0.642,1.4853,0.1629, ...] | [0.3181,0.4221,0.1823, ...] | 3.227 | | ||
|
|
||
| ## Column Description | ||
|
|
||
| - **id**: | ||
| A unique identifier for each RNA sequence entry. | ||
|
|
||
| - **design**: | ||
| The name given to each RNA design by contributors, used for easy reference. | ||
|
|
||
| - **sequence**: | ||
| The nucleotide sequence of the RNA molecule, represented using the standard RNA bases: | ||
|
|
||
| - **A**: Adenine | ||
| - **C**: Cytosine | ||
| - **G**: Guanine | ||
| - **U**: Uracil | ||
|
|
||
| - **secondary_structure**: | ||
| The secondary structure of the RNA represented in dot-bracket notation, using up to three types of symbols to indicate base pairing and unpaired regions, as per bpRNA's standard: | ||
|
|
||
| - **Dots (`.`)**: Represent unpaired nucleotides. | ||
| - **Parentheses (`(` and `)`)**: Represent base pairs in standard stems (page 1). | ||
| - **Square Brackets (`[` and `]`)**: Represent base pairs in pseudoknots (page 2). | ||
| - **Curly Braces (`{` and `}`)**: Represent base pairs in additional pseudoknots (page 3). | ||
|
|
||
| - **reactivity**: | ||
| A list of normalized reactivity values for each nucleotide, representing the likelihood that a nucleotide is unpaired. | ||
| High reactivity indicates high flexibility (unpaired regions), and low reactivity corresponds to paired or structured regions. | ||
|
|
||
| - **errors**: | ||
| Arrays of floating-point numbers indicating the experimental errors corresponding to the measurements in the **reactivity**. | ||
| These values help quantify the uncertainty in the degradation rates and reactivity measurements. | ||
|
|
||
| - **signal_to_noise**: | ||
| The signal-to-noise ratio calculated from the reactivity and error values, providing a measure of data quality. | ||
|
|
||
| ## Related Datasets | ||
|
|
||
| - [eternabench-switch](https://huggingface.co/datasets/multimolecule/eternabench-switch) | ||
| - [eternabench-external.1200](https://huggingface.co/datasets/multimolecule/eternabench-external.1200): EternaBench-External dataset with maximum sequence length of 1200 nucleotides. | ||
|
|
||
| ## License | ||
|
|
||
| This dataset is licensed under the [AGPL-3.0 License](https://www.gnu.org/licenses/agpl-3.0.html). | ||
|
|
||
| ```spdx | ||
| SPDX-License-Identifier: AGPL-3.0-or-later | ||
| ``` | ||
|
|
||
| ## Citation | ||
|
|
||
| ```bibtex | ||
| @article{waymentsteele2022rna, | ||
| author = {Wayment-Steele, Hannah K and Kladwang, Wipapat and Strom, Alexandra I and Lee, Jeehyung and Treuille, Adrien and Becka, Alex and {Eterna Participants} and Das, Rhiju}, | ||
| journal = {Nature Methods}, | ||
| month = oct, | ||
| number = 10, | ||
| pages = {1234--1242}, | ||
| publisher = {Springer Science and Business Media LLC}, | ||
| title = {{RNA} secondary structure packages evaluated and improved by high-throughput experiments}, | ||
| volume = 19, | ||
| year = 2022 | ||
| } | ||
| ``` |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,63 @@ | ||
| # MultiMolecule | ||
| # Copyright (C) 2024-Present MultiMolecule | ||
|
|
||
| # This program is free software: you can redistribute it and/or modify | ||
| # it under the terms of the GNU Affero General Public License as published by | ||
| # the Free Software Foundation, either version 3 of the License, or | ||
| # any later version. | ||
|
|
||
| # This program is distributed in the hope that it will be useful, | ||
| # but WITHOUT ANY WARRANTY; without even the implied warranty of | ||
| # MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the | ||
| # GNU Affero General Public License for more details. | ||
|
|
||
| # You should have received a copy of the GNU Affero General Public License | ||
| # along with this program. If not, see <http://www.gnu.org/licenses/>. | ||
|
|
||
| from __future__ import annotations | ||
|
|
||
| import os | ||
|
|
||
| import danling as dl | ||
| import pandas as pd | ||
| import torch | ||
|
|
||
| from multimolecule.datasets.conversion_utils import ConvertConfig as ConvertConfig_ | ||
| from multimolecule.datasets.conversion_utils import save_dataset | ||
|
|
||
| torch.manual_seed(1016) | ||
|
|
||
| cols = [ | ||
| "id", | ||
| "design", | ||
| "sequence", | ||
| "secondary_structure", | ||
| "reactivity", | ||
| "errors", | ||
| "signal_to_noise", | ||
| ] | ||
|
|
||
|
|
||
| def convert_dataset_(df: pd.DataFrame): | ||
| df.signal_to_noise = df.signal_to_noise.str.split(":").str[-1].astype(float) | ||
| df = df.rename(columns={"ID": "id", "design_name": "design", "structure": "secondary_structure"}) | ||
| df = df.sort_values("id") | ||
| df = df[cols] | ||
| return df | ||
|
|
||
|
|
||
| def convert_dataset(convert_config): | ||
| train = dl.load_pandas(convert_config.train_path) | ||
| test = dl.load_pandas(convert_config.test_path) | ||
| save_dataset(convert_config, {"train": convert_dataset_(train), "test": convert_dataset_(test)}) | ||
|
|
||
|
|
||
| class ConvertConfig(ConvertConfig_): | ||
| root: str = os.path.dirname(__file__) | ||
| output_path: str = os.path.basename(os.path.dirname(__file__)).replace("_", "-") | ||
|
|
||
|
|
||
| if __name__ == "__main__": | ||
| config = ConvertConfig() | ||
| config.parse() # type: ignore[attr-defined] | ||
| convert_dataset(config) |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,117 @@ | ||
| --- | ||
| language: rna | ||
| tags: | ||
| - Biology | ||
| - RNA | ||
| license: | ||
| - agpl-3.0 | ||
| size_categories: | ||
| - 1K<n<10K | ||
| task_categories: | ||
| - text-generation | ||
| - fill-mask | ||
| task_ids: | ||
| - language-modeling | ||
| - masked-language-modeling | ||
| pretty_name: EternaBench-External | ||
| library_name: multimolecule | ||
| --- | ||
|
|
||
| # EternaBench-External | ||
|
|
||
|  | ||
|
|
||
| EternaBench-External consists of 31 independent RNA datasets from various biological sources, including viral genomes, mRNAs, and synthetic RNAs. | ||
| These sequences were probed using techniques such as SHAPE-CE, SHAPE-MaP, and DMS-MaP-seq to understand RNA secondary structures under different experimental and biological conditions. | ||
| This dataset serves as a benchmark for evaluating RNA structure prediction models, with a particular focus on generalization to natural RNA molecules. | ||
|
|
||
| ## Disclaimer | ||
|
|
||
| This is an UNOFFICIAL release of the [EternaBench-External](https://github.com/eternagame/EternaBench) by Hannah K. Wayment-Steele, et al. | ||
|
|
||
| **The team releasing EternaBench-External did not write this dataset card for this dataset so this dataset card has been written by the MultiMolecule team.** | ||
|
|
||
| ## Dataset Description | ||
|
|
||
| - **Homepage**: https://multimolecule.danling.org/datasets/eternabench_external | ||
| - **Point of Contact**: [Rhiju Das](https://biochemistry.stanford.edu/people/rhiju-das/) | ||
|
|
||
| This dataset includes RNA sequences from various biological origins, including viral genomes and mRNAs, and covers a wide range of probing methods like SHAPE-CE and icSHAPE. | ||
| Each dataset entry provides sequence information, reactivity profiles, and RNA secondary structure data. | ||
| This dataset can be used to examine how RNA structures vary under different conditions and to validate structural predictions for diverse RNA types. | ||
|
|
||
| ## Example Entry | ||
|
|
||
| | name | sequence | reactivity | seqpos | class | dataset | | ||
| | ----------------------------------------------------------- | ----------------------- | -------------------------------- | -------------------- | ---------- | -------------- | | ||
| | Dadonaite,2019 Influenza genome SHAPE(1M7) SSII-Mn(2+) Mut. | TTTACCCACAGCTGTGAATT... | [0.639309,0.813297,0.622869,...] | [7425,7426,7427,...] | viral_gRNA | Dadonaite,2019 | | ||
|
|
||
| ## Column Description | ||
|
|
||
| - **name**: | ||
| The name of the dataset entry, typically including the experimental setup and biological source. | ||
|
|
||
| - **sequence**: | ||
| The nucleotide sequence of the RNA molecule, represented using the standard RNA bases: | ||
|
|
||
| - **A**: Adenine | ||
| - **C**: Cytosine | ||
| - **G**: Guanine | ||
| - **U**: Uracil | ||
|
|
||
| - **reactivity**: | ||
| A list of normalized reactivity values for each nucleotide, representing the likelihood that a nucleotide is unpaired. | ||
| High reactivity indicates high flexibility (unpaired regions), and low reactivity corresponds to paired or structured regions. | ||
|
|
||
| - **seqpos**: | ||
| A list of sequence positions corresponding to each nucleotide in the **sequence**. | ||
|
|
||
| - **class**: | ||
| The type of RNA sequence, can be one of the following: | ||
|
|
||
| - SARS-CoV-2_gRNA | ||
| - mRNA | ||
| - rRNA | ||
| - small RNAs | ||
| - viral_gRNA | ||
|
|
||
| - **dataset**: | ||
| The source or reference for the dataset entry, indicating its origin. | ||
|
|
||
| ## Variations | ||
|
|
||
| This dataset is available in four variants: | ||
|
|
||
| - [eternabench-external.1200](https://huggingface.co/datasets/multimolecule/eternabench-external.1200): EternaBench-External dataset with maximum sequence length of 1200 nucleotides. | ||
| - [eternabench-external.900](https://huggingface.co/datasets/multimolecule/eternabench-external.900): EternaBench-External dataset with maximum sequence length of 900 nucleotides. | ||
| - [eternabench-external.600](https://huggingface.co/datasets/multimolecule/eternabench-external.600): EternaBench-External dataset with maximum sequence length of 600 nucleotides. | ||
| - [eternabench-external.300](https://huggingface.co/datasets/multimolecule/eternabench-external.300): EternaBench-External dataset with maximum sequence length of 300 nucleotides. | ||
|
|
||
| ## Related Datasets | ||
|
|
||
| - [eternabench-cm](https://huggingface.co/datasets/multimolecule/eternabench-cm) | ||
| - [eternabench-switch](https://huggingface.co/datasets/multimolecule/eternabench-switch) | ||
|
|
||
| ## License | ||
|
|
||
| This dataset is licensed under the [AGPL-3.0 License](https://www.gnu.org/licenses/agpl-3.0.html). | ||
|
|
||
| ```spdx | ||
| SPDX-License-Identifier: AGPL-3.0-or-later | ||
| ``` | ||
|
|
||
| ## Citation | ||
|
|
||
| ```bibtex | ||
| @article{waymentsteele2022rna, | ||
| author = {Wayment-Steele, Hannah K and Kladwang, Wipapat and Strom, Alexandra I and Lee, Jeehyung and Treuille, Adrien and Becka, Alex and {Eterna Participants} and Das, Rhiju}, | ||
| journal = {Nature Methods}, | ||
| month = oct, | ||
| number = 10, | ||
| pages = {1234--1242}, | ||
| publisher = {Springer Science and Business Media LLC}, | ||
| title = {{RNA} secondary structure packages evaluated and improved by high-throughput experiments}, | ||
| volume = 19, | ||
| year = 2022 | ||
| } | ||
| ``` |
Oops, something went wrong.