Check out our MetaCerberus ReadTheDocs Documentation and Tutorial!
MetaCerberus transforms raw sequencing (i.e. genomic, transcriptomics, metagenomics, metatranscriptomic) data into knowledge. It is a start to finish python code for versatile analysis of the Functional Ontology Assignments for Metagenomes (FOAM), KEGG, CAZy/dbCAN, VOG, pVOG, PHROG, COG, and a variety of other databases including user customized databases via Hidden Markov Models (HMM) for functional annotation for complete metabolic analysis across the tree of life (i.e., bacteria, archaea, phage, viruses, eukaryotes, and whole ecosystems). MetaCerberus also provides automatic differential statistics using DESeq2/EdgeR, pathway enrichments with GAGE, and pathway visualization with Pathview R.
 Art by Andra Buchan
Art by Andra Buchan
- Mamba install from bioconda with all dependencies:
- Install mamba using conda
conda install mamba- NOTE: Make sure you install mamba in your base conda environment unless you have OSX with ARM architecture (M1/M2 Macs). Follow the OSX-ARM instructions below if you have a Mac with ARM architecture.
- Install MetaCerberus with mamba
mamba create -n metacerberus -c conda-forge -c bioconda metacerberus
conda activate metacerberus
metacerberus.py --setup
metacerberus.py --download- Set up conda environment
conda create -y -n metacerberus
conda activate metacerberus
conda config --env --set subdir osx-64- Install mamba, python, and pydantic inside the environment
conda install -y -c conda-forge mamba python=3.10 "pydantic<2"- Install MetaCerberus with mamba
mamba install -y -c conda-forge -c bioconda metacerberus
metacerberus.py --setup
metacerberus.py --download- NOTE: Mamba is the fastest installer. Anaconda or miniconda can be slow. Also, install mamba from conda not from pip. The pip mamba doesn't work for install.
- Anaconda install from bioconda with all dependencies:
conda create -n metacerberus -c conda-forge -c bioconda metacerberus -y
conda activate metacerberus
metacerberus.py --setup
metacerberus.py --download
git clone https://github.com/raw-lab/MetaCerberus.git
cd MetaCerberus
bash install_metacerberus.sh
conda activate MetaCerberus
metacerberus.py --downloadWe also have a lite version of MetaCerberus on anaconda that only depends on the very basic dependencies.
This can make it a bit faster and easier to install as it is less likely to have conflicts with other dependencies on the system.
To install the "lite" version, use "metacerberus-lite" instead of "metacerberus" from Bioconda, following the details listed above.
mamba create -n metacerberus -c conda-forge -c bioconda metacerberus-lite
conda activate metacerberus
metacerberus.py --setup
metacerberus.py --downloadAdditional dependencies such as fastqc and fastp can be installed in the environment manually if desired for those steps in the pipeline.
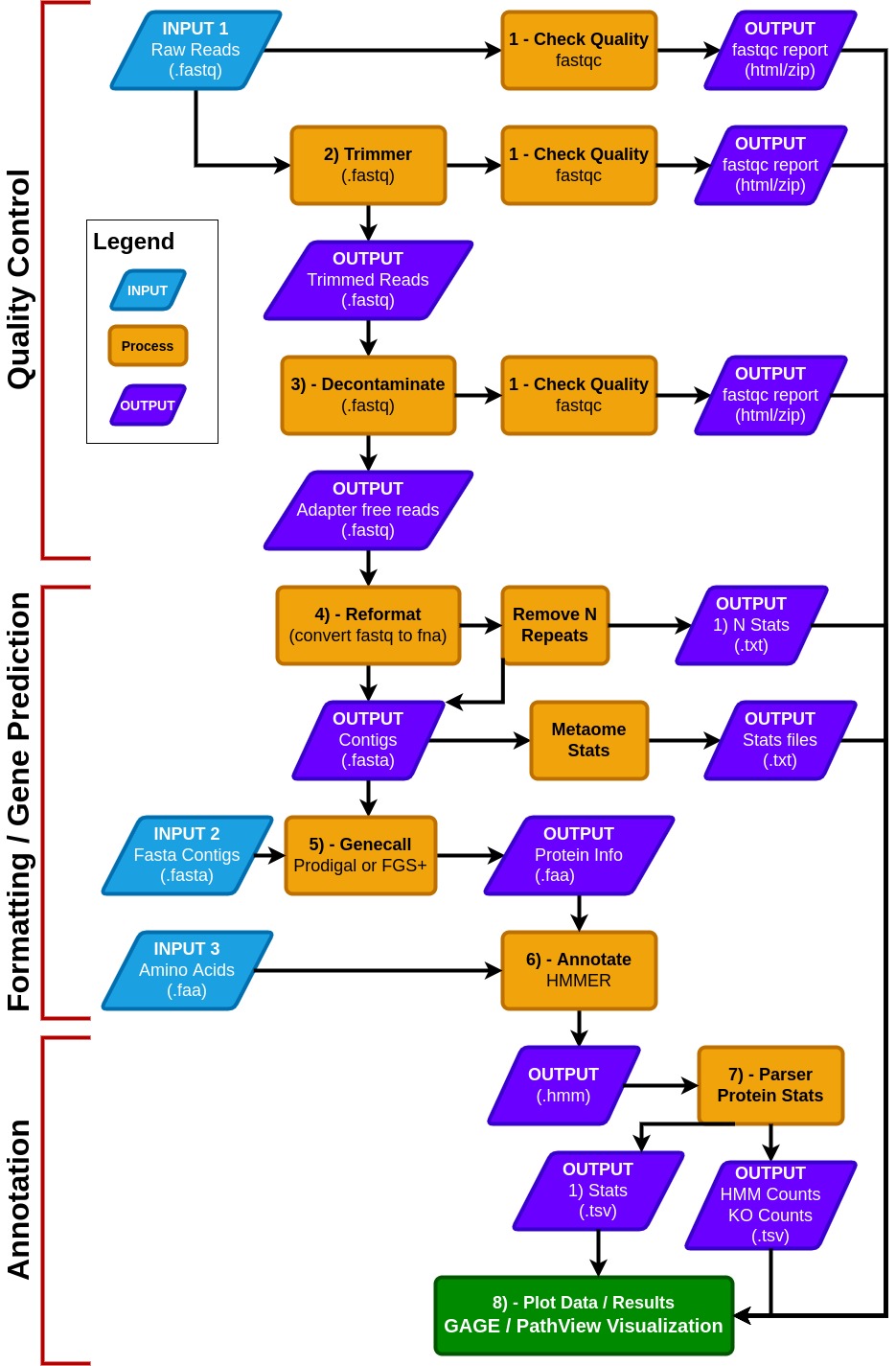
MetaCerberus has three basic modes: quality control (QC) for raw reads, formatting/gene prediction, and annotation.
- MetaCerberus can use three different input files: 1) raw read data from any sequencing platform (Illumina, PacBio, or Oxford Nanopore), 2) assembled contigs, as MAGs, vMAGs, isolate genomes, or a collection of contigs, 3) amino acid fasta (.faa), previously called pORFs.
- We offer customization, including running all databases together, individually or specifying select databases. For example, if a user wants to run prokaryotic or eukaryotic-specific KOfams, or an individual database alone such as dbCAN, both are easily customized within MetaCerberus.
- In QC mode, raw reads are quality controlled via FastQC prior and post trim FastQC. Raw reads are then trimmed via data type; if the data is Illumina or PacBio, fastp is called, otherwise it assumes the data is Oxford Nanopore then Porechop is utilized PoreChop.
- If Illumina reads are utilized, an optional bbmap step to remove the phiX174 genome is available or user provided contaminate genome. Phage phiX174 is a common contaminant within the Illumina platform as their library spike-in control. We highly recommend this removal if viral analysis is conducted, as it would provide false positives to ssDNA microviruses within a sample.
- We include a --skip_decon option to skip the filtration of phiX174, which may remove common k-mers that are shared in ssDNA phages.
- In the formatting and gene prediction stage, contigs and genomes are checked for N repeats. These N repeats are removed by default.
- We impute contig/genome statistics (e.g., N50, N90, max contig) via our custom module Metaome Stats.
- Contigs can be converted to pORFs using Prodigal, FragGeneScanRs, and Prodigal-gv as specified by user preference.
- Scaffold annotation is not recommended due to N's providing ambiguous annotation.
- Both Prodigal and FragGeneScanRs can be used via our --super option, and we recommend using FragGeneScanRs for samples rich in eukaryotes.
- FragGeneScanRs found more ORFs and KOs than Prodigal for a stimulated eukaryote rich metagenome. HMMER searches against the above databases via user specified bitscore and e-values or our minimum defaults (i.e., bitscore = 25, e-value = 1 x 10-9 ).
- From any NextGen sequencing technology (from Illumina, PacBio, Oxford Nanopore)
- type 1 raw reads (.fastq format)
- type 2 nucleotide fasta (.fasta, .fa, .fna, .ffn format), assembled raw reads into contigs
- type 3 protein fasta (.faa format), assembled contigs which genes are converted to amino acid sequence
- If an output directory is given, that folder will be created where all files are stored.
- If no output directory is specified, the 'results_metacerberus' subfolder will be created in the current directory.
- Gage/Pathview R analysis provided as separate scripts within R.
- We use Plotly to visualize the data
- Once the program is executed the html reports with the visuals will be saved to the last step of the pipeline.
- The HTML files require plotly.js to be present. One has been provided in the package and is saved to the report folder.
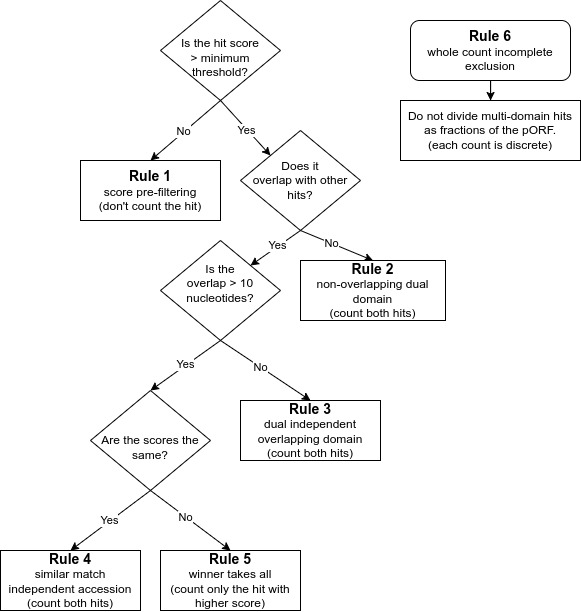
- Rule 1 is for finding high quality matches across databases. It is a score pre-filtering module for pORFs thresholds: which states that each pORF match to an HMM is recorded by default or a user-selected cut-off (i.e., e-value/bit scores) per database independently, or across all default databases (e.g, finding best hit), or per user specification of the selected database.
- Rule 2 is to avoid missing genes encoding proteins with dual domains that are not overlapping. It is imputed for non-overlapping dual domain module pORF threshold: if two HMM hits are non-overlapping from the same database, both are counted as long as they are within the default or user selected score (i.e., e-value/bit scores).
- Rule 3 is to ensure overlapping dual domains are not missed. This is the dual independent overlapping domain module for convergent binary domain pORFs. If two domains within a pORF are overlapping <10 amino acids (e.g, COG1 and COG4) then both domains are counted and reported due to the dual domain issue within a single pORF. If a function hits multiple pathways within an accession, both are counted, in pathway roll-up, as many proteins function in multiple pathways.
- Rule 4 is the equal match counter to avoid missing high quality matches within the same protein. This is an independent accession module for a single pORF: if both hits within the same database have equal values for both e-value and bit score but are different accessions from the same database (e.g., KO1 and KO3) then both are reported.
- Rule 5 is the ‘winner take all’ match rule for providing the best match. It is computed as the winner takes all module for overlapping pORFs: if two HMM hits are overlapping (>10 amino acids) from the same database the lowest resulting e-value and highest bit score wins.
- Rule 6 is to avoid partial or fractional hits being counted. This ensures that only whole discrete integer counting (e.g., 0, 1, 2 to n) are computed and that partial or fractional counting is excluded.
conda activate metacerberus
metacerberus.py --prodigal lambda.fna --hmm ALL --dir_out lambda_dirconda activate metacerberus
metacerberus.py --prodigal lambda.fna --hmm KOFam_all --dir_out lambda_ko-only_dirconda activate metacerberus
metacerberus.py --prodigal ecoli.fna --hmm KOFam_prokaryote --dir_out ecoli_ko-only_dirconda activate metacerberus
metacerberus.py --fraggenescan human.fna --hmm KOFam_eukaryote --dir_out human_ko-only_dirconda activate metacerberus
metacerberus.py --prodigal lambda.fna --hmm VOG, PHROG --dir_out lambda_vir-only_dir- NOTE: You can pick any single database you want for your analysis including KOFam_all, COG, VOG, PHROG, CAZy or specific KO databases for eukaryotes and prokaryotes (KOFam_eukaryote or KOFam_prokaryote).
conda activate metacerberus
metacerberus.py --prodigal lambda.fna --hmm Custom.hmm --dir_out lambda_vir-only_dirconda activate metacerberus
metacerberus.py --prodigal [input_folder] --illumina --meta --dir_out [out_folder] conda activate metacerberus
metacerberus.py --fraggenescan [input_folder] --illumina --meta --dir_out [out_folder] conda activate metacerberus
metacerberus.py --prodigal [input_folder] --nanopore --meta --dir_out [out_folder]conda activate metacerberus
metacerberus.py --fraggenescan [input_folder] --nanopore --meta --dir_out [out_folder] conda activate metacerberus
metacerberus.py --prodigal [input_folder] --pacbio --meta --dir_out [out_folder]conda activate metacerberus
metacerberus.py --fraggenescan [input_folder] --pacbio --meta --dir_out [out_folder]conda activate metacerberus
metacerberus.py --super [input_folder] --pacbio/--nanopore/--illumina --meta --dir_out [out_folder]- Note: Fraggenescan will work for prokaryotes and viruses/bacteriophage but prodigal will not work well for eukaryotes.
- python >= 3.8
| Tool | Version | Publication |
|---|---|---|
| Fastqc | 0.12.1 | None |
| Fastp | 0.23.4 | Chen et al. 2018 |
| Porechop | 0.2.4 | None |
| bbmap | 39.06 | None |
| Prodigal | 2.6.3 | Hyatt et al. 2010 |
| FragGeneScanRs | v1.1.0 | Van der Jeugt et al. 2022 |
| Prodigal-gv | 2.2.1 | Camargo et al. 2023 |
| Phanotate | 1.5.0 | McNair et al. 2019 |
| HMMER | 3.4 | Johnson et al. 2010 |
| HydraMPP | 0.0.4 | None |
All pre-formatted databases are present at OSF
| Database | Last Update | Version | Publication | MetaCerberus Update Version |
|---|---|---|---|---|
| KEGG/KOfams | 2024-01-01 | Jan24 | Aramaki et al. 2020 | beta |
| FOAM/KOfams | 2017 | 1 | Prestat et al. 2014 | beta |
| COG | 2020 | 2020 | Galperin et al. 2020 | beta |
| dbCAN/CAZy | 2023-08-02 | 12 | Yin et al., 2012 | beta |
| VOG | 2024-10-06 | 225 | Website | 1.4 |
| pVOG | 2016 | 2016 | Grazziotin et al. 2017 | 1.2 |
| PHROG | 2022-06-15 | 4 | Terizan et al., 2021 | 1.2 |
| PFAM | 2023-09-12 | 36 | Mistry et al. 2020 | 1.3 |
| TIGRfams | 2018-06-19 | 15 | Haft et al. 2003 | 1.3 |
| PGAPfams | 2023-12-21 | 14 | Tatusova et al. 2016 | 1.3 |
| AMRFinder-fams | 2024-02-05 | 2024-02-05 | Feldgarden et al. 2021 | 1.3 |
| NFixDB | 2024-01-22 | 2 | Bellanger et al. 2024 | 1.3 |
| GVDB | 2021 | 1 | Aylward et al. 2021 | 1.3 |
| Pads Arsenal | 2019-09-09 | 1 | Zhang et al. 2020 | Coming soon |
| efam-XC | 2021-05-21 | 1 | Zayed et al. 2021 | Coming soon |
| NMPFams | 2021 | 1 | Baltoumas et al. 2024 | Coming soon |
| MEROPS | 2017 | 1 | Rawlings et al. 2018 | Coming soon |
| FESNov | 2024 | 1 | Rodríguez del Río et al. 2024 | Coming soon |
- NOTE: The KEGG database contains KOs related to Human disease. It is possible that these will show up in the results, even when analyzing microbes. eggNOG and FunGene database are coming soon. If you want a custom HMM build please let us know by email or leaving an issue.
To run a custom database, you need a HMM containing the protein family of interest and a metadata sheet describing the HMM required for look-up tables and downstream analysis. For the metadata information you need an ID that matches the HMM and a function or hierarchy. See example below.
| ID | Function |
|---|---|
| HMM1 | Sugarase |
| HMM2 | Coffease |
- If the metacerberus environment is not used, make sure the dependencies are in PATH or specified in the config file.
- Run metacerberus.py with the options required for your project.
usage: metacerberus.py [--setup] [--update] [--list-db] [--download [DOWNLOAD ...]] [--uninstall] [-c CONFIG] [--prodigal PRODIGAL [PRODIGAL ...]]
[--fraggenescan FRAGGENESCAN [FRAGGENESCAN ...]] [--super SUPER [SUPER ...]] [--prodigalgv PRODIGALGV [PRODIGALGV ...]]
[--phanotate PHANOTATE [PHANOTATE ...]] [--protein PROTEIN [PROTEIN ...]] [--hmmer-tsv HMMER_TSV [HMMER_TSV ...]] [--class CLASS]
[--illumina | --nanopore | --pacbio] [--dir-out DIR_OUT] [--replace] [--keep] [--hmm HMM [HMM ...]] [--db-path DB_PATH] [--address ADDRESS]
[--port PORT] [--meta] [--scaffolds] [--minscore MINSCORE] [--evalue EVALUE] [--remove-n-repeats] [--skip-decon] [--skip-pca] [--cpus CPUS]
[--chunker CHUNKER] [--grouped] [--version] [-h] [--adapters ADAPTERS] [--qc_seq QC_SEQ]
Setup arguments:
--setup Setup additional dependencies [False]
--update Update downloaded databases [False]
--list-db List available and downloaded databases [False]
--download [DOWNLOAD ...]
Downloads selected HMMs. Use the option --list-db for a list of available databases, default is to download all available databases
--uninstall Remove downloaded databases and FragGeneScan+ [False]
Input files
At least one sequence is required.
accepted formats: [.fastq, .fq, .fasta, .fa, .fna, .ffn, .faa]
Example:
> metacerberus.py --prodigal file1.fasta
> metacerberus.py --config file.config
*Note: If a sequence is given in [.fastq, .fq] format, one of --nanopore, --illumina, or --pacbio is required.:
-c CONFIG, --config CONFIG
Path to config file, command line takes priority
--prodigal PRODIGAL [PRODIGAL ...]
Prokaryote nucleotide sequence (includes microbes, bacteriophage)
--fraggenescan FRAGGENESCAN [FRAGGENESCAN ...]
Eukaryote nucleotide sequence (includes other viruses, works all around for everything)
--super SUPER [SUPER ...]
Run sequence in both --prodigal and --fraggenescan modes
--prodigalgv PRODIGALGV [PRODIGALGV ...]
Giant virus nucleotide sequence
--phanotate PHANOTATE [PHANOTATE ...]
Phage sequence (EXPERIMENTAL)
--protein PROTEIN [PROTEIN ...], --amino PROTEIN [PROTEIN ...]
Protein Amino Acid sequence
--hmmer-tsv HMMER_TSV [HMMER_TSV ...]
Annotations tsv file from HMMER (experimental)
--class CLASS path to a tsv file which has class information for the samples. If this file is included scripts will be included to run Pathview in R
--illumina Specifies that the given FASTQ files are from Illumina
--nanopore Specifies that the given FASTQ files are from Nanopore
--pacbio Specifies that the given FASTQ files are from PacBio
Output options:
--dir-out DIR_OUT path to output directory, defaults to "results-metacerberus" in current directory. [./results-metacerberus]
--replace Flag to replace existing files. [False]
--keep Flag to keep temporary files. [False]
Database options:
--hmm HMM [HMM ...] A list of databases for HMMER. 'ALL' uses all downloaded databases. Use the option --list-db for a list of available databases [KOFam_all]
--db-path DB_PATH Path to folder of databases [Default: under the library path of MetaCerberus]
MPP options:
--address ADDRESS Address for distributed MPP. local=no networking, host=make this machine a host, ip-address=connect to remote host [local]
--port PORT The port to listen/connect to [24515]
optional arguments:
--meta Metagenomic nucleotide sequences (for prodigal) [False]
--scaffolds Sequences are treated as scaffolds [False]
--minscore MINSCORE Score cutoff for parsing HMMER results [60]
--evalue EVALUE E-value cutoff for parsing HMMER results [1e-09]
--remove-n-repeats Remove N repeats, splitting contigs [False]
--skip-decon Skip decontamination step [False]
--skip-pca Skip PCA [False]
--cpus CPUS Number of CPUs to use per task. System will try to detect available CPUs if not specified [Auto Detect]
--chunker CHUNKER Split files into smaller chunks, in Megabytes [Disabled by default]
--grouped Group multiple fasta files into a single file before processing. When used with chunker can improve speed
--version, -v show the version number and exit
-h, --help show this help message and exit
--adapters ADAPTERS FASTA File containing adapter sequences for trimming
--qc_seq QC_SEQ FASTA File containing control sequences for decontamination
Args that start with '--' can also be set in a config file (specified via -c). Config file syntax allows: key=value, flag=true, stuff=[a,b,c] (for details, see syntax at
https://goo.gl/R74nmi). In general, command-line values override config file values which override defaults.| File Extension | Description Summary | MetaCerberus Update Version |
|---|---|---|
| .gff | General Feature Format | 1.3 |
| .gbk | GenBank Format | 1.3 |
| .fna | Nucleotide FASTA file of the input contig sequences. | 1.3 |
| .faa | Protein FASTA file of the translated CDS/ORFs sequences. | 1.3 |
| .ffn | FASTA Feature Nucleotide file, the Nucleotide sequence of translated CDS/ORFs. | 1.3 |
| .html | Summary statistics and/or visualizations, in step 10 folder | 1.3 |
| .txt | Statistics relating to the annotated features found. | 1.3 |
| level.tsv | Various levels of hierachical steps that is tab-separated file from various databases | 1.3 |
| rollup.tsv | All levels of hierachical steps that is tab-separated file from various databases | 1.3 |
| .tsv | Final Annotation summary, Tab-separated file of all features from various databases | 1.3 |
After processing the HMM files MetaCerberus calculates a KO (KEGG Orthology) counts table from KEGG/FOAM for processing through GAGE and PathView. GAGE is recommended for pathway enrichment followed by PathView for visualize the metabolic pathways. A "class" file is required through the --class option to run this analysis. As we are unsure which comparisons you want to make thus you have to make a class.tsv so the code will know the comparisons you want to make.
| Sample | Class |
|---|---|
| 1A | rhizobium |
| 1B | non-rhizobium |
The output is saved under the step_10-visualizeData/combined/pathview folder. Also, at least 4 samples need to be used for this type of analysis.
GAGE and PathView also require internet access to be able to download information from a database. MetaCerberus will save a bash script 'run_pathview.sh' in the step_10-visualizeData/combined/pathview directory along with the KO Counts tsv files and the class file for running manualy in case MetaCerberus was run on a cluster without access to the internet.
MetaCerberus uses HydraMPP for distributed processing. This is compatible with both multiprocessing on a single node (computer) or multiple nodes in a cluster.
MetaCerberus has been tested on a cluster using Slurm https://github.com/SchedMD/slurm.
*note the extra flag "--hydraMPP-slurm $SLURM_JOB_NODELIST" when running MetaCerberus. HydraMPP uses this to setup the SLURM jobs.
sbatch example_script.shexample script:
#!/usr/bin/env bash
#SBATCH --job-name=test-job
#SBATCH --nodes=3
#SBATCH --tasks-per-node=1
#SBATCH --cpus-per-task=16
#SBATCH --mem=128MB
#SBATCH -e slurm-%j.err
#SBATCH -o slurm-%j.out
#SBATCH --mail-type=END,FAIL,REQUEUE
echo "====================================================="
echo "Start Time : $(date)"
echo "Job ID/Name : $SLURM_JOBID / $SLURM_JOB_NAME"
echo "Node List : $SLURM_JOB_NODELIST"
echo "Num Tasks : $SLURM_NTASKS total [$SLURM_NNODES nodes @ $SLURM_CPUS_ON_NODE CPUs/node]"
echo "======================================================"
echo ""
# Load any modules or resources here
conda activate MetaCerberus
# run MetaCerberus
metacerberus.py --prodigal [input_folder] --illumina --dir_out [out_folder] --hydraMPP-slurm $SLURM_JOB_NODELISTBoth edgeR and DeSeq2 R have the highest sensitivity when compared to other algorithms that control type-I error when the FDR was at or below 0.1. EdgeR and DESeq2 all perform fairly well in simulation and via data splitting (so no parametric assumptions). Typical benchmarks will show limma having stronger FDR control across all types of datasets (it’s hard to beat the moderated t-test), and edgeR and DESeq2 having higher sensitivity for low counts (makes sense as limma has to filter these out / down-weight them to use the normal model on log counts). Further information about type I errors are present from Mike Love's vignette here.
MetaCerberus as a community resource as recently acquired FunGene, we welcome contributions of other experts expanding annotation of all domains of life (viruses, bacteria, archaea, eukaryotes). Please send us an issue on our MetaCerberus GitHub open an issue; or email us we will fully annotate your genome, add suggested pathways/metabolisms of interest, make custom HMMs to be added to MetaCerberus and FunGene.
This is copyrighted by University of North Carolina at Charlotte, Jose L Figueroa III, Eliza Dhungal, Madeline Bellanger, Cory R Brouwer and Richard Allen White III. All rights reserved. MetaCerberus is a bioinformatic tool that can be distributed freely for academic use only. Please contact us for commerical use. The software is provided “as is” and the copyright owners or contributors are not liable for any direct, indirect, incidental, special, or consequential damages including but not limited to, procurement of goods or services, loss of use, data or profits arising in any way out of the use of this software.
If you are publishing results obtained using MetaCerberus, please cite:
Figueroa III JL, Dhungel E, Bellanger M, Brouwer CR, White III RA. 2024.
MetaCerberus: distributed highly parallelized HMM-based processing for robust functional annotation across the tree of life. Bioinformatics
Figueroa III JL, Dhungel E, Brouwer CR, White III RA. 2023.
MetaCerberus: distributed highly parallelized HMM-based processing for robust functional annotation across the tree of life. bioRxiv
The informatics point-of-contact for this project is Dr. Richard Allen White III.
If you have any questions or feedback, please feel free to get in touch by email.
Dr. Richard Allen White III
Jose Luis Figueroa III
Or open an issue.