-
Notifications
You must be signed in to change notification settings - Fork 0
/
in_Tutorial03LabSetUp.Rmd
428 lines (256 loc) · 18.5 KB
/
in_Tutorial03LabSetUp.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
---
title: "Tutorial 3: Setting up each lab"
toc_depth: 4
---
```{r setup, include=FALSE}
# OPTIONS -----------------------------------------------
knitr::opts_chunk$set(echo = TRUE,
warning=FALSE,
message = FALSE)
```
```{r, include=FALSE}
# PACKAGES-----------------------------------------------
# Tutorial packages
library(vembedr)
library(skimr)
library(yarrr)
library(RColorBrewer)
library(GGally)
library(tidyverse)
library(plotly)
library(readxl)
library(rvest)
library(biscale)
library(tidycensus)
library(cowplot)
library(units)
```
In this tutorial we are going to cover setting up each lab. This involves making projects, packages and R Markdown documents.
# Starting a new lab
## About Projects
In this class, we will be using R-Projects:
- On Rstudio cloud, this is how you set up each lab.
- On the desktop, this is a folder that will store everything to do with each lab in one place on your computer. Each lab will be its own project.
This is incredibly useful - it means that if you switch from R-Cloud, to the lab computers, to your laptop, all you have to do is to move the folder and everything will just work. Equally, its easy to compare labs.
Learn more here.
[](https://www.linkedin.com/learning/learning-the-r-tidyverse/why-should-you-use-projects-in-rstudio?u=76811570)
We will cover how to make projects for R studio cloud and desktop in the set up below.
## Projects using Desktop
Everything should now be installed. If not, go back to Tutorial 2.
```{r,Tut_Fig10, echo=FALSE, out.width="80%",fig.align='center'}
knitr::include_graphics('./Figures/fig10_Startup_Logos.png')
```
1. IMPORTANT! If you haven't already, on an easy to access place on your computer, make a folder called GEOG364. This is where ALL your labs are going to live. <br>
2. Now everything is installed, open R-studio (NOT R!). <br>
Steps 3-7 are shown graphically in the figure below.
3. Go to the Main File menu at the [very top left]{.underline} and click `New Project` <br>\
4. Select `New Directory`, then `New Project`<br>\
5. Name your project *GEOG364-Lab1-PROJECT* (or whatever lab it us) <br>\
6. Under "create project as a subdirectory of", hit the browse button and find your GEOG364 folder (you just need to be in the folder, you don't need to have selected anything). Press open <br>\
7. Finally, press `Create Project`<br>
```{r, Tut_Fig11, echo=FALSE}
knitr::include_graphics('./Figures/fig11_projectcreation.png')
```
### How to check you are in a project {.unnumbered}
R will change slightly. If you look at the top of the screen in the title bar, it should say something like *GEOG364-Lab1-PROJECT - R Studio*.
The Files tab should have gone to your project folder. Essentially, R-Studio is now "looking" inside your Lab 1 folder, making it easier to find your data and output your results.
```{r, Tut_Fig12, echo=FALSE, fig.cap="How to check you are in a project"}
knitr::include_graphics('./Figures/fig12_projectcheck.png')
```
<br>
If you want one, final check, try typing this into the console (INCLUDING THE EMPTY PARANTHESES/BRACKETS), press enter and see if it prints out the location of Lab 1 on your computer. If not, talk to an instructor.
```{r, eval=FALSE}
getwd()
```
<br><br>
### Returning to your lab project
OK, let's imagine that you get halfway through your lab and your computer dies. How do you get back to your Lab work? Try this now. Close down R-Studio.
To reopen a lab:
1. **DO NOT RE-OPEN R-STUDIO!**
2. Instead navigate on your computer to your *GEOG364/GEOG364-Lab1-PROJECT* folder.
3. Double click on the *GEOG364-Lab1-PROJECT*.RProj file.
This will reopen R for that specific lab, so you can continue where you left off. It means you can also open several versions of R studio for multiple labs, which can be very useful in staying sane
```{r, Tut_Fig13, echo=FALSE}
knitr::include_graphics('./Figures/fig13_projectopen.png')
```
<br><br>
## Projects using Rstudio/posit Cloud
Go to this webpage. <https://login.posit.cloud/login> You can get here from any web-browser, You do not need a special computer.
When you log in, you should see a dashboard like this.
```{r, Tut_Fig14, echo=FALSE, fig.cap = "*The Cloud dashboard*",fig.align='center'}
knitr::include_graphics('./Figures/fig14_clouddash.png')
```
YOU SHOULD MAKE A NEW PROJECT FOR EACH LAB.
You make a new project by clicking the button on the top right. You can open different projects in different tabs on your browser. You can also go back to the work-space at any time.
### Returning to your lab project
On the cloud this is very easy, just go back to <https://posit.cloud/content/yours>
### Downloading files or switching to desktop
Let's say you want to switch to the desktop but save your progress. Or you want to submit your work to Canvas!
To switch to desktop, download/install the desktop version using the tutorials here. Then make a new project using the instructions above.
Either way:
1. On the R-Studio Cloud website; inside your project, go to the files tab (next to Projects/Help in one quadrant. You will see a list of files, one with the file type .Rmd (your code) and one with .html(the website you made when you pressed knit). Look at the red circle in the pic below
2. Click the check-box to the left of the .RmD file
3. Look just above at the Files quadrant settings menu. Click the blue "more" cogwheel icon. (see diagram). You might need to make R-Studio full screen to see it, it cuts off if the window is too small.
4. Now click Export This will download the file
5. REPEAT FOR THE HTML (you can do them together but it exports as a zip)
6. Submit both files on Canvas or put them in your project file.
```{r, Tut_Fig15, echo=FALSE, fig.cap = "*Downloading the files*",fig.align='center'}
knitr::include_graphics('./Figures/fig15_export.png')
```
### Uploading files {.unnumbered}
Sometimes you want to put your code into R-Studio Cloud, for example if for one week you don't want to bring your laptop.
Make a project for the lab on the cloud as above, then click the `upload` button inside your project in the files tab and upload your files. You will only need the .RmD file and any data files.
```{r, Tut_Fig16, echo=FALSE, fig.cap = "This is zoomed in to your files quadrant",fig.align='center'}
knitr::include_graphics('./Figures/fig16_import.png')
```
# The RStudio screen
You will be greeted by three panels:
- The interactive R console (entire left)
- Environment/History (tabbed in upper right)
- Files/Plots/Packages/Help/Viewer (tabbed in lower right)
```{r, Tut_Fig17, echo=FALSE, fig.align='center'}
knitr::include_graphics('./Figures/fig17_quadrants.png')
```
If you wish to learn more about what these windows do, have a look at this resource, from the Pirates Guide to R: <https://bookdown.org/ndphillips/YaRrr/the-four-rstudio-windows.html>.
If you have used R before, you might see that there are variables and plots etc already loaded. It is always good to clear these before you start a new analysis. To do this, click the little broom symbol in your environment tab. <br><br>
# Changing global options
Some of the default R settings are worth changing to make life smoother. We do this in the main global options menu.
You only have to do this once! Follow the arrows in this figure (sections explained below).
```{r, Tut_Fig18, echo=FALSE, fig.align='center'}
knitr::include_graphics('./Figures/fig18_global.png')
```
There are two places you might find this menu:
1. Click on the tools menu button on the VERY top right of the screen,-\> Global Options OR.. <br>
2. Click on the R-studio menu button on the top left of the screen, then click Preferences.<br><br>
## Main options screen
On the main options screen (see figure above), I suggest:
- UNCLICK "Restore most recently opened project at startup"
- UNCLICK "Restore .RData into workspace on startup"
- Set "Save workspace to .RData on" exit to Never
- UNCLICK "Restore previously open source documents on startup"
Note, yours will probably show a different default working directory.
<br><br>
## Changing the appearance
OPTIONAL: You can also click the appearances menu item to change how the screen looks (see the figure above). For example, maybe you concentrate better with a dark background or with pink font.<br><br>
## Moving the windows around
OPTIONAL: Like me, you might find you like the different quadrants in a different order. To change this, look at the Panes layout menu item (you can also get here via the `View` menu,then `/Panes/Pane Layout`) I tend to like the console to be top left and scripts to be top right, with the plots and environment on the bottom - but this is personal choice.<br>
<br><br>
# R-Packages/libraries
<br>
## What are packages?
As described earlier, we program in R by typing a series of commands. R is open source and anyone can create a new command. Over the last 20 years,tens of millions of new custom commands have been created.
Commands tend to be grouped together into collections called `Packages` or `Libraries` (two names for the same thing). For example, one package contains the complete works of Shakespeare; another allows interactive website design; another allows advanced Bayesian statistics. There is a package for literally everything and there are now over 20,000 packages available. You can see a selection here: <https://cran.r-project.org/web/packages/available_packages_by_name.html> <br>
**A close analogy is your phone:** There are millions of apps available from banking, to 50 different calendar apps. You don't have every app in the world installed on your phone - and you don't have every app you *do* download running at the same time. Instead you download the apps that you think you will need (occasionally downloading a new one on the fly) - and when you need to use an app, you click on it to open.<br>
This is exactly the same in R. To access the commands in a package we need these two steps:
1. ONCE ONLY: Download the package from the internet, in the way you download an app from the app store
2. EVERY TIME: Load the packages you want, like clicking an app icon to open it. <br><br>
## Downloading a new package
### Manually click
This is like going to the app store to get a new app. Just like you only go to the app store once, this is a one-off for each package. NOTE! For R studio cloud online, you might have to do this for each project.
- Look for the quadrant with the packages tab in it.
- You will see a list of packages/apps that have already been installed.
- Click the INSTALL button in the Packages tab menu (on the left)
- Start typing the package name and it will show up (check the include dependencies box). Install the package.
### Little yellow banner
- R will sometime tell you that you are missing a package (sometimes a little yellow ribbon), click yes to install!
```{r, Fig19, echo=FALSE, fig.align='center',out.width="100%"}
knitr::include_graphics('./Figures/fig19_yellowbanner.png')
```
### Using the console
You can also use a command to install a new package:
```{r, eval=FALSE}
install.packages("bardr")
```
**DO NOT INCLUDE ANY INSTALL.PACKAGES COMMANDS IN YOUR FINAL SCRIPT. ALWAYS TYPE/RUN THIS COMMAND IN THE CONSOLE WINDOW.** The command means "go to the app store" and download a package. You don't want to do this when pressing knit. <br><br>
### PRACTICE FOR YOU
Please install the rmdformats package onto your computer/the cloud. You will need this in the nest step.
### Problem solving
#### Why isn't my package downloading? Its frozen
Sometimes R will ask you if you want to install binaries or other things. IT WILL ASK YOU THIS QUESTION THROUGH "SPEAKING" IN THE CONSOLE. It expects you to type yes or no, and to press enter to continue. Try yes, if it doesn't work (esp xfun), try no. <br><br>
#### R keeps asking to restart.
Sometimes R-Studio might want to restart when downloading packages and occasionally gets confused. If it keeps asking, press cancel, close R-studio, reopen and try again. If it really doesn't want to work, open the R program itself and run in the console there (or talk to Dr G) <br><br>
## Loading/Using the commands inside a package
Just as going to the app store doesn't check your credit-card balance or make an Instagram post, simply *downloading* a package from the app-store doesn't make the commands immediately available. For that you need to load it (just as you click on a phone app to open it).
**I will tell you which packages you need for each lab, but if R tells you it wants a package, then install it AND load it.**
This can be done with the `library()` command.
For example, this command loads the full works of Shakespeare from the the bardr package. (<https://www.rdocumentation.org/packages/bardr/versions/0.0.9>). If you want to try this, you might need to first install bardr using the instructions above.
```{r, eval=FALSE}
library(bardr)
```
<br>
### Using a single command from a package
ADVANCED: Sometimes several packages name a command the same word and you want to specify which package you want to use.
You can do this using the :: symbol. For example, this command *forces* the computer to use the 'dplyr package' version of filter.
```{r, eval=FALSE}
dplyr::filter(mydata)
```
<br><br>
### Problem Solving
#### Nothing happened!
If you have managed to load a package successfully, often nothing happens - this is great! It means it loaded the package without errors.
#### There was a load of text output - an error?
Hard to tell. So I suggest running the library command TWICE! This is because many packages will print "friendly messages" or "welcome text" the first time you load them.
For example, this is what shows up when you install the tidyverse package. The welcome text is indicating the sub-packages that tidyverse downloaded and also that some commands now have a different meaning.
```{r, fig20, echo=FALSE, fig.cap = "Tidyverse install messages",fig.align='center',out.width="100%"}
knitr::include_graphics('./Figures/fig20_friendlytext.png')
```
**To find out if what you are seeing is a friendly message or an error, run the command again. If you run it a second time and and nothing happens, then you're fine.**
<br>
# Creating a Markdown document
This will be your lab report. Remind yourself of what Rmarkdown is using Tutorial 1, here <https://rmarkdown.rstudio.com>, or via this short video
```{r,echo=FALSE,fig.align='center'}
embed_url("https://vimeo.com/178485416?embedded=true&source=video_title&owner=22717988")
```
## Templates
In the past, you used to have to know a lot of computer commands to get markdown documents to work. Every month it seems to get easier, and there are now a load of professional templates available.
The ones we are going to use are from the package `rmdformats`. Have a look at the templates here: <https://juba.github.io/rmdformats/>. If none of these appeal, google RMarkdown templates for more.
Install the package `rmdformats` using the packages tutorial above.
## Making your Markdown file
Go to the File menu on the top left, then click New File - **R-Markdown**. If this is your first time ever, it might ask to download some packages to be able to do this. Say yes. Eventually a window will open up:
```{r, fig21a, echo=FALSE,fig.align='center',out.width="80%"}
knitr::include_graphics('./Figures/fig21a_Markdown.png')
```
Click "From Template" and **select a template of your choice**. Click here to see what different templates look like:
<https://juba.github.io/rmdformats/>
```{r, fig21b, echo=FALSE,fig.align='center'}
knitr::include_graphics('./Figures/fig21B_Markdown templates.png')
```
If you don't see any templates, you needed to have installed the rmdformats package (see above). There are many packages containing different templates. You can see which package each template belongs to below (red). You should also be running your project - so it should auto save your file to your lab 1 folder.
It will ask you to name your file. Give it a relevant name including your email ID, like Lab-2_hlg5155. Press OK.
```{r, fig21c, echo=FALSE,fig.align='center',out.width="80%"}
knitr::include_graphics('./Figures/fig21c_Creating doc.png')
```
<br> A new file should appear on your screen.
```{r, fig21d, echo=FALSE,fig.align='center',out.width="80%"}
knitr::include_graphics('./Figures/fig21d_Markdown.png')
```
Essentially, we have some space for text, some space for code, and a space at the top of the file where we can add information about themes/styles etc called "YAML".
Each grey code area is called a "code chunk". To run the code inside it, click the little green arrow on the top right.
Depending on the template, your file might contain some friendly text to explain what is going on. Read through it, then you are welcome to delete it from the after the lower --- onwards.
```{r, fig21e, echo=FALSE,fig.align='center'}
knitr::include_graphics('./Figures/fig21e_Markdown.png')
```
## THREE IMPORTANT BUTTONS - READ THIS
## Visual mode
It is MUCH easier to edit markdown documents in the new visual mode. Essentially instead of having to remember text short cuts like \* for bold, you can edit the text part as though you were using a word processor. NOTE HEADERS where it says "Normal", this allows you to make auto tables of contents.
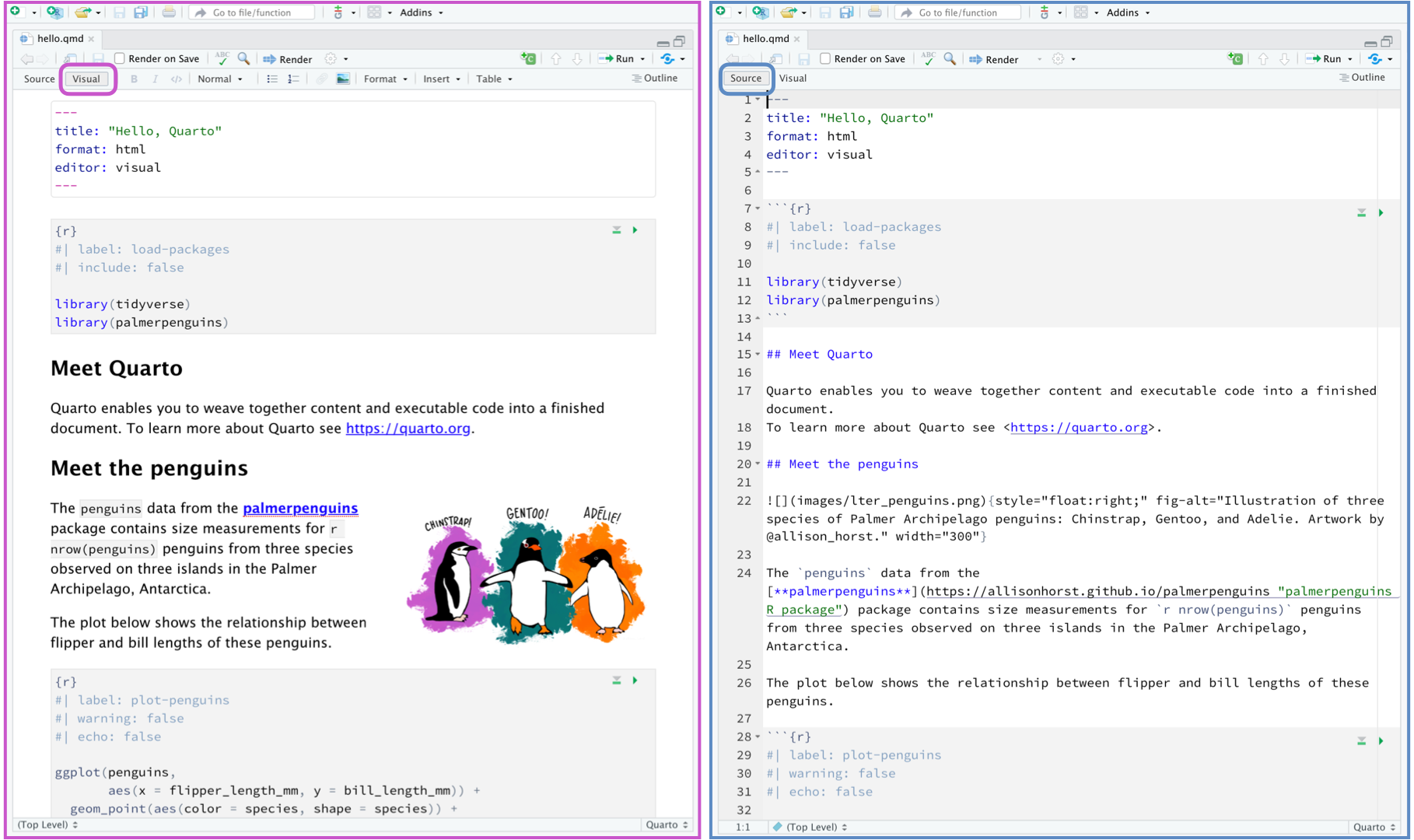
### Creating a new code chunk.
Code chunks are mini consoles where you can run R commands. We will talk more about them in the next tutorial.
```{r, Fig22, echo=FALSE,fig.align='center'}
knitr::include_graphics('./Figures/fig22_CodeChunk.png')
```
On the top right there are a suite of buttons for adding a new code chunk, running code etc. The green one adds a new code chunk. To run an individual code chunk you will press the green arrow on its top right e.g.
```{r, Fig23 ,echo=FALSE,fig.align='center'}
knitr::include_graphics('./Figures/fig23_RunCodeChunk.png')
```
### Knitting
```{r, fig24, echo=FALSE,fig.align='center'}
knitr::include_graphics('./Figures/fig24_knit.png')
```
The file on your screen isn't the finished article. To see how it will look as a final version, we need to "knit" it. Go to the top of the .Rmd file, find the `knit` button. Press it (you might have to first save your script if you haven't already, then press it again)
You should see that the Markdown tab "builds" your document and you get an output as a website. The html should also be saved into your project folder.
For example, from my other class, here is a file with markdown and knitted output.
```{r, fig25, echo=FALSE,fig.align='center'}
knitr::include_graphics('./Figures/fig25_AllMarkdownElements.png')
```
<br> <br>