-
Notifications
You must be signed in to change notification settings - Fork 3
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Refactor training and inference to make it feasible for unit tests #26
Comments
|
This overall looks like a sensible structure. I am missing some information regarding what the individual function do and how you plan to set up testing to better understand the restructuring though. Some questions in this regard:
|
|
Thanks for the feedback @Wiebke . To address each comment individually:
Yes, this function is supposed to cover the pydantic validation of model-related parameters.
Good point, in the diagram above those parameters are captured in the
These will be in separate functions in the new design layout below.
Based on my initial glance, most of them will share the same return object, so putting the entire function in utilities should be fine. For those that may need specifications, the plan is to break into even smaller functions and only put common parts as utility functions.
I will do another round of check for names, during development I will also include docstrings.
In training, the loop will occur in the |
|
As suggested by @dylanmcreynolds and @Wiebke , here is a new version of flow chat that follows the sequential order of functions: flowchart TD
subgraph Training
classDef utils fill:#f96
Begin[/yaml_path\]
A("load_params(yaml_path)"\n '''load all params from yaml file'''):::utils
Begin --> A
B("validate_io_params(params)"\n '''pydantic validation of io params'''):::utils
A --> B
C("initialize_tiled_dataset(clients)"\n '''construct TiledDataset class'''):::utils
B --> C
D("build_qlty_object(qlty_params)"\n '''build qlty object'''):::utils
C --> D
E("prepare_dataset(tiled_dataset)" \n '''extract image and mask array''')
D --> E
F("normalization(images)" \n '''min-max normalization of images''')
E --> F
G("array_to_tensor(array)" \n '''transform normalized images and mask arrays to tensors''')
F --> G
H("qlty_cropping(images, masks, qlty_obj)" \n '''perform qlty cropping and construct TensorDataset''')
G --> H
I("construct_dataloader(training_dataset)" \n '''split and convert into train_loader, val_loader''')
H --> I
J("get_model_params(network_name, params) \n '''pydantic validation of params specific to models"):::utils
I --> J
K("build_network(model_params)"\n '''build dlsia algorithm with params''')
J --> K
L("find_device()"\n '''find either gpu or cpu'''):::utils
K --> L
M("define_criterion(criterion_name, weights, device)"\n '''build criterion with given class weights''')
L --> M
N("train_network(network, train_loader, val_loader, optimizer, device, criterion)"\n '''train network using dlsia built-in Trainer class''')
M --> N
O("save_trained_network(trained_network, model_dir)"\n '''create local directory and save trained model''')
N --> O
End[\trained network saved in local dir/]
O --> End
end
flowchart TD
subgraph Inference
classDef utils fill:#f96
Begin[/yaml_path\]
A("load_params(yaml_path)"\n '''load all params from yaml file'''):::utils
Begin --> A
B("validate_io_params(params)"\n '''pydantic validation of io params'''):::utils
A --> B
C("initialize_tiled_dataset(clients)"\n '''construct TiledDataset class'''):::utils
B --> C
D("allocate_array_space(result_client)"\n '''pre-allocate chunk in tiled client for result saving''')
C --> D
E("build_qlty_object(qlty_params)"\n '''build qlty object'''):::utils
D --> E
F("get_model_params(network_name, params) \n '''pydantic validation of params specific to models"):::utils
E --> F
G("find_device()"\n '''find either gpu or cpu'''):::utils
F --> G
H("load_network(model_dir)"\n '''load trained model from local directory''')
G --> H
I("extract_slice(tiled_dataset)" \n '''extract a single image array for segmentation''')
H --> I
J("normalization(images)" \n '''min-max normalization of the slice''')
I --> J
K("array_to_tensor(array)" \n '''transform normalized image array to tensor''')
J --> K
L("qlty_cropping(images, masks, qlty_obj)" \n '''perform qlty cropping and construct TensorDataset'''):::utils
K --> L
M("construct_dataloader(slice)" \n '''pass TensorDataset to inference_loader''')
L --> M
N("segment(inference_loader)"\n '''segment all patches for a single slice''')
M --> N
O("stitch(qlty_obj, result_array)"\n '''stitch back from segmented patches to original image''')
N --> O
P("tiled_client.write_block(seg_result, block=(frame_idx, 0, 0))"\n '''save single slice back to tiled''')
O --> P
P --> I
End[\inference results saved in tiled server/]
P --> End
end
|
|
Thanks for the updates. This is very nice. Just curious, @TibbersHao , what does the orange indicate for some of the boxes? For training, I'd suggest combining |
Those stand for common functions shared between both scripts and will be put into the utility.
Sounds good to me. |
|
The diagrams look great so far. Just a couple of extra suggestions:
I was also thinking if we'd like to add the partial inference step directly to the train script to avoid some duplicated actions that increase processing time, such as trained model loading, data loading, cropping, etc. I think the waiting time for the training process has not been an issue so far, but this could become problematic when using previous (maybe large) segmented results for re-training/fine-tuning processes followed by partial inference. |
|
Thanks for the feedback @taxe10 , here are my thoughts:
For tracking, do you mean leaving a log message when transfer has been completed, or there are more to be done? Also for the training part, since the moving is happening within the DLSIA trainer class, I do not have a good way in my mind to handle this, other than making some changes to DLSIA itself. For inference it should be more straightforward.
Yes good catch. Originally I thought the normalization for training takes the percentile of the whole training stack, while inference only takes the percentile of that single slice, so they are different. I checked the code again, it appears to be the same code, so yes, they will be in utility functions.
I was planning to have the argmax step combined within the stitch function, for clarity reason of the flow chart (it's already too long). Yes we won't miss any logical steps in the original code, thanks for checking.
I like this idea, as I don't see in any scenario which we won't run partial inference after training, especially in model tuning phase. What do other people think @Wiebke @dylanmcreynolds @ahexemer ? My only concern so far is that the frontend will likely make some adjustments to accompany this change. |
|
I think it makes sense to do the quicker inference in connection with training and am not too concerned about this requiring changes in the front-end as we are maintaining both. However, we need to ensure that it is generally possible to segment a subset of the data even in the absence of a mask, such that users loading a previously trained model have the option to try segmentation on select slices. |
|
I agree; we need to run inference after training on all slices containing any labeling. There is no need for a front-end change. I also agree with Wiebke that we need to allow the loading of trained models and segment a subset/all slices. Inference, as we learned the hard way, needs to run on multi GPU/nodes if at all possible. |
|
Just a heads up that @Giselleu is currently working on adding multi GPU support to the inference step in this pipeline. My initial suggestion was to make changes to the current version of the algorithm such that these 2 efforts can be completed in parallel, but it'd be great for you both to coordinate this work. |
|
Multi GPU support for inference would be amazing - please let me know how
you think to implement this. I was thinking myself about a dask-type setup,
but have not yet had the time to dive into this. Especially with the 3D
ensemble networks being able to push this across a couple of cards (within
a notebook) is very handy. This goes hand in hand with the qlty chunker
options of course. The 2D qlty i have right now is ok, but I need to make a
new version that mimics that qlty3DLarge method, that uses zarr arrays as
intermediate cache when performing stitching operations across spatial
dimensions.
P
…On Thu, Jun 27, 2024 at 3:14 PM Tanny Chavez Esparza < ***@***.***> wrote:
Just a heads up that @Giselleu <https://github.com/Giselleu> is currently
working on adding multi GPU support to the inference step in this pipeline.
My initial suggestion was to make changes to the current version of the
algorithm such that these 2 efforts can be completed in parallel, but it'd
be great for you both to coordinate this work.
—
Reply to this email directly, view it on GitHub
<#26 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ADWIEE7XGFYFMUGTGMGN52DZJRXEFAVCNFSM6AAAAABJQ4WYP2VHI2DSMVQWIX3LMV43OSLTON2WKQ3PNVWWK3TUHMZDCOJVGU4TKOJZGA>
.
You are receiving this because you are subscribed to this thread.Message
ID: <mlexchange/mlex_dlsia_segmentation_prototype/issues/26/2195595990@
github.com>
--
------------------------------------------------------------------------------------------
Peter Zwart
Staff Scientist, Molecular Biophysics and Integrated Bioimaging
Berkeley Synchrotron Infrared Structural Biology
Biosciences Lead, Center for Advanced Mathematics for Energy Research
Applications
Lawrence Berkeley National Laboratories
1 Cyclotron Road, Berkeley, CA-94703, USA
Cell: 510 289 9246
------------------------------------------------------------------------------------------
|
|
I plan to try torch DistributedDataParallel approach for implementing Multi-node Multi-GPU inference supported by Nersc Perlmutter's GPU resources. This will distribute the data across GPUs when defining the Dataloader, create replica of models on each GPU and synchronize gradients (during training). I am new to the segmentation app and code, will need to see how the current inference and training architecture works. |
The current layout of training and inference scripts contain lengthy functions which covers the whole process in one step, this is not friendly to write unit tests to boost the robustness. Thus refactoring big functions to small and testable functions is needed for future development.
To propose a new training script (
train.py) without changing current logic:Note: Individual functions have been wrapped in rectangle, common utility functions which will be called in both training and inference are colored in orange.
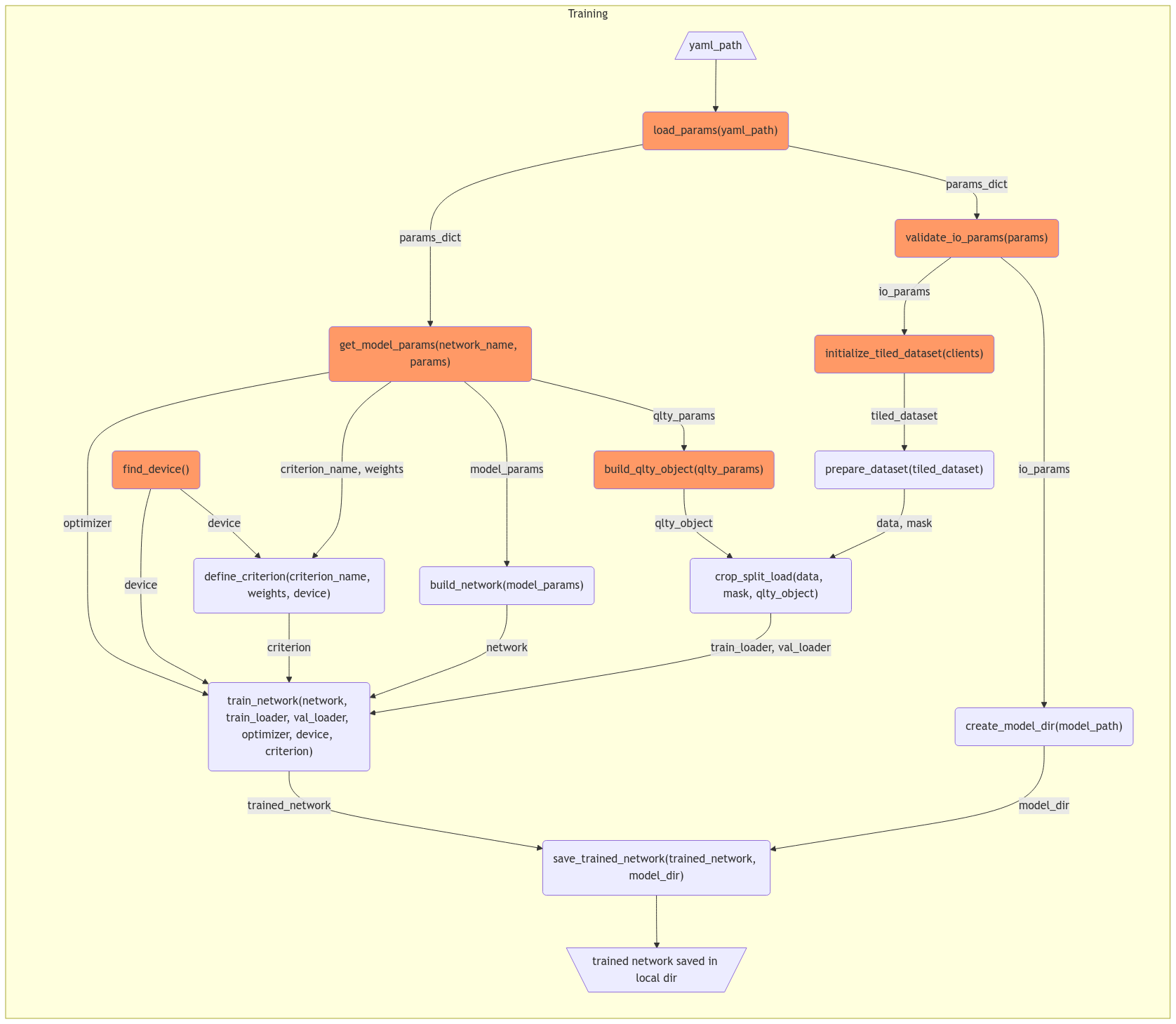
To propose a new inference script (
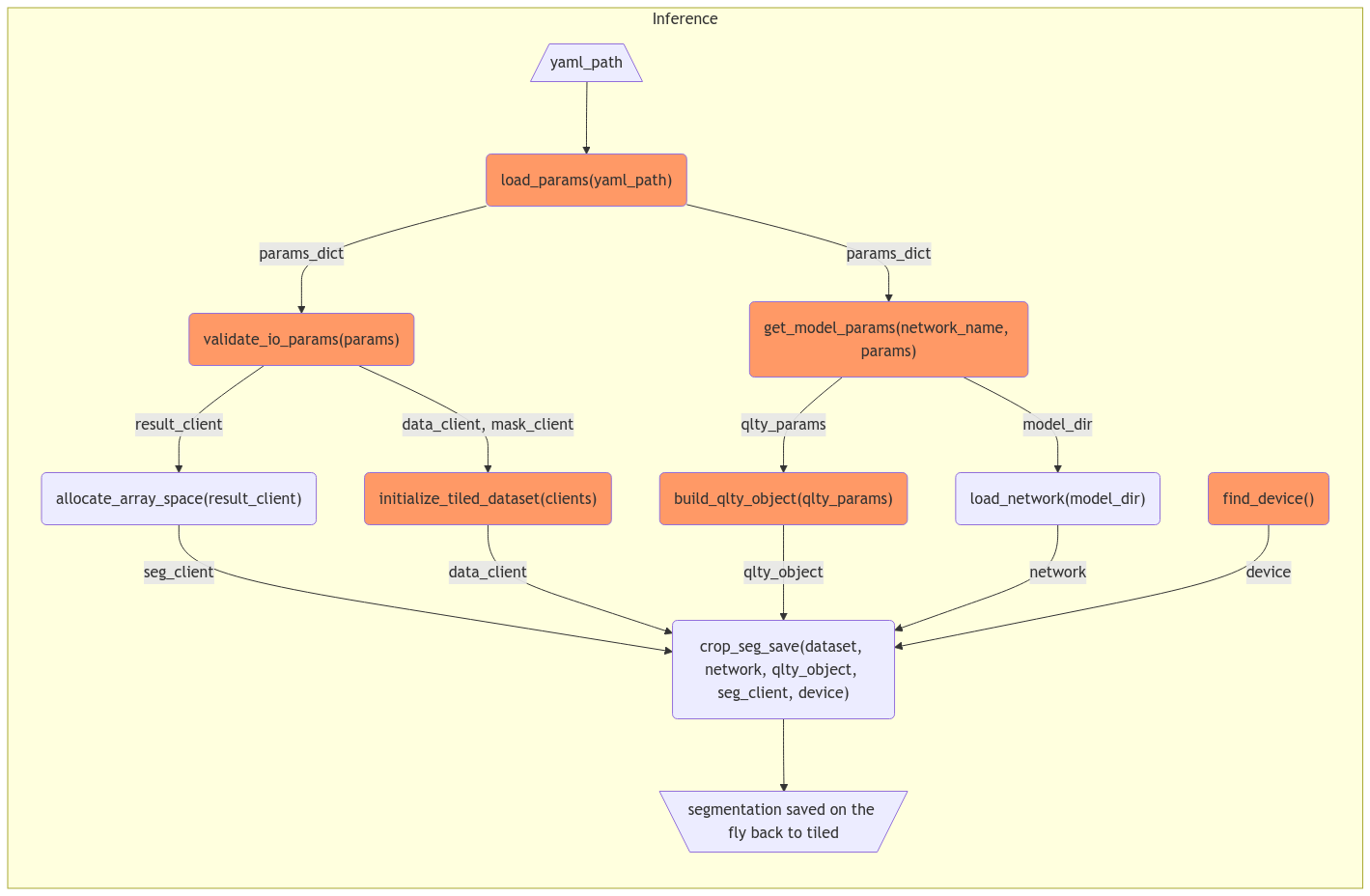
segment.py) without changing current logic:Feedbacks are welcomed @dylanmcreynolds @Wiebke @taxe10 @ahexemer.
The text was updated successfully, but these errors were encountered: