Maintained by Marco A. Lopez-Sanchez - This website was last modified: 2024/02/07
GrainSizeTools is a free, open-source, cross-platform script written in Python that provides tools for (1) describing and visualizing grain size populations, (2) estimating differential stress for different mineral phases via paleopizometers and (3) apply stereology methods to approximate the actual distribution (3D) of grain size from sections. The script does not require previous experience with the Python programming language (see the documentation below and FAQ). For users with coding skills, the script is organized in a modular (functional) way facilitating the reuse and code extension.
Latest release: v3.0.2
Date: 2020/12/31
See notes at https://github.com/marcoalopez/GrainSizeTools/releases/tag/v3.0.2
Features at a glance
Import and manipulate tabular-like datasets including txt, csv, or excel formats.
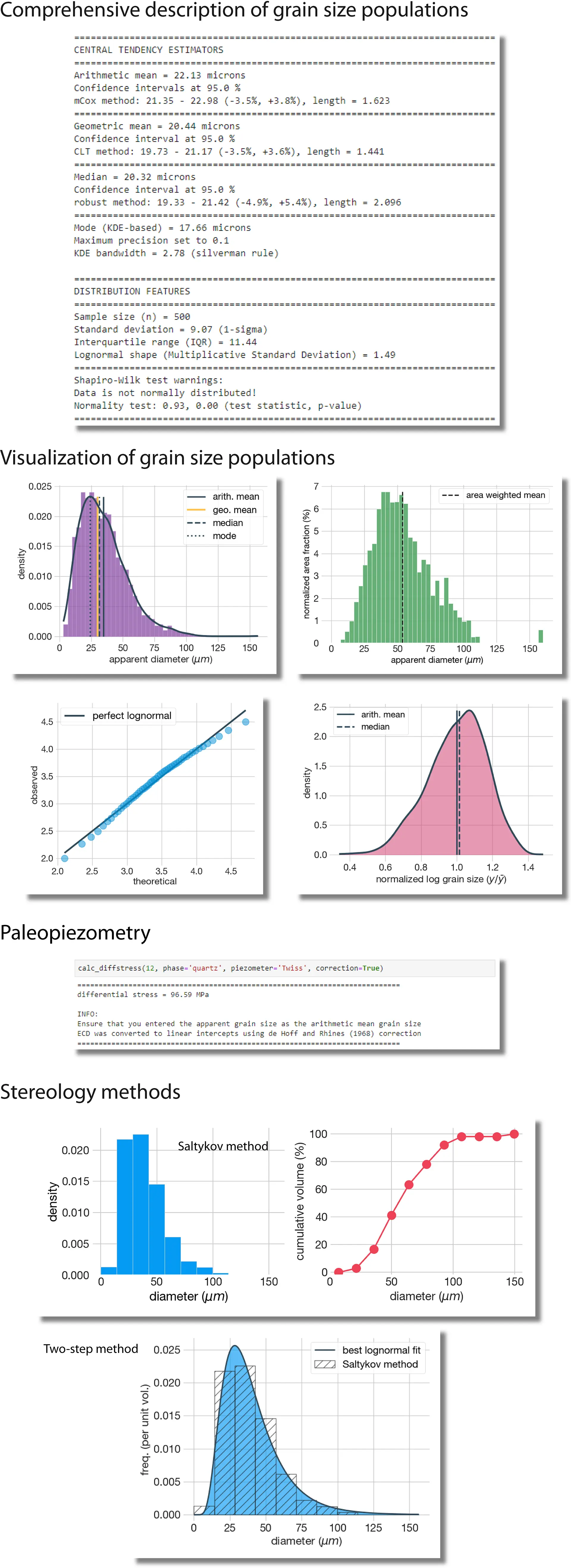
Full automatic descriptive statistics of grain size populations including:
Several averages (arithmetic and geometric means, median, and frequency peak ("mode") via Gaussian Kernel Density Estimator)
Estimation of robust confidence intervals (including some specific methods for lognormal populations such as the modified Cox or the GCI method)
Measures of dispersion and shape of the population
Normality and lognormality test
Estimate differential stress using paleopiezometers. It includes multiple piezometric relations for quartz, olivine, calcite, and feldspar (others planned!)
Ready-to-publish plots in bitmap or vector format (see screenshots below), including:
Histograms and kernel density estimates
Area- or volume-weighted plots
Normalized plots
Quantile-quantile plots, and more
Stereology methods (approximate the actual 3D grain size distribution from data collected in plane sections):
Saltykov method
Two-step method (lognormal fitting, shape characterization)
Download
You can download the script at the following sites:
https://github.com/marcoalopez/GrainSizeTools/releases (primary source)
http://figshare.com/articles/GrainSizeTools_script/1383130
https://sourceforge.net/projects/grainsizetools/
Documentation
Others
Screenshots (v3.0+)
Citation guidelines
If you have utilized the script, I kindly request that you consider citing the following paper:
Lopez-Sanchez, Marco A. (2018). GrainSizeTools: a Python script for grain size analysis and paleopiezometry based on grain size. Journal of Open Source Software, 3(30), 863, https://doi.org/10.21105/joss.00863
By acknowledging this paper, you provide proper credit to the author and acknowledge his work.
License
GrainSizeTools script is licensed under the Apache License, Version 2.0 (the "License")
The documentation of GrainSizeTools script is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (CC BY-NC-SA 4.0).
Community guidelines
The GitHub website hosting the project provides several options (you will need a GitHub account, it’s free!):
Open a discussion: This is a place to:
Ask questions you are wondering about.
Share ideas.
Engage with the developers (still just me).
Open and issue: This is a place to track bugs or requests for specific features on the scripts.
Create a pull request: You modified, corrected or added a feature to one of the notebooks and send it for one of the developers to review it and add it to the main page.
Copyright © 2017-2024 Marco A. Lopez-Sanchez
Warning
Information presented on this website and the documentation of the script is provided without any express or implied warranty and may include technical inaccuracies or typing errors; the author reserve the right to modify or enhance the content of this website as well as the documentation of the script at any time without previous notice. This webpage and the documentation is not liable for the content of external links.
Hosted on GitHub Pages — This website was created with Typora