You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
Merge module supports multiple scenarios and keys management is the "key" here. Consider following scenario where we have a two selectors to compare specific AVAL level from the same dataset across different factor values - for example AVAL increase between two visits
In above example, PARAMCD and AVISITN in both selectors have just one value which means that it's no longer the key for these ANLs as they were filtered-out (does not distinguish the rows). The reason to drop these keys is that it wouldn't be possible if by = c("USUBJID", "STUDYID", "PARAMCD", "AVISITN") because they have a different values and it's not possible to merge them together.
For scda data we have no problem with this as we have a static data and keys are relevant for our test data. Problems starts when SMEs create their own apps which are using real data. The keys used to be different than default teal keys, but the keys follow GDSR requirements. For example ADLB keys are STUDYID USUBJID PARAMCD BASETYPE AVISITN ATPTN DTYPE ADTM LBSEQ ASPID vs teal STUDYID USUBJID PARAMCD AVISIT. SMEs require the change in the keys to fit GDSR standards which complicates the design little bit.
If we go back to the last example which is using ADLB. Extending set of keys will result in joining by keys which have not been filtered-out. This means that ANLs will be joined by "STUDYID", "USUBJID", "ATPTN", "DTYPE", "ADTM", "LBSEQ", "ASPID" if we put only PARAMCD and AVISITN as a filter_spec.
Current apps which are created with PARAMCD and/or AVISITfilter_spec might fail because output ANL will be merged on a columns which have a different values across the subjects. One can run above code and see that the result of inner_join is empty data.frame. Also our problem is that GDSR keys are not the same what we think they are. Some of the keys in the datasets does not make any row unique adding them - which means that for us they are just values. For example AVISITN is a number of a patient visit while ADTM can be a data of the visit. This means that adding ADTM to the keys does not make any row unique, because STUDYID, USUBJID, PARAMCD and AVISITN make them unique already.
Problem description
Design of data_extract/data_merge is as follows. Consider columns taken from datasets, filtered separately and merged all together.

Above can be express in the following code
Merge module supports multiple scenarios and keys management is the "key" here. Consider following scenario where we have a two selectors to compare specific AVAL level from the same dataset across different factor values - for example AVAL increase between two visits
In above example, PARAMCD and AVISITN in both selectors have just one value which means that it's no longer the key for these ANLs as they were filtered-out (does not distinguish the rows). The reason to drop these keys is that it wouldn't be possible if
by = c("USUBJID", "STUDYID", "PARAMCD", "AVISITN")because they have a different values and it's not possible to merge them together.For
scdadata we have no problem with this as we have a static data and keys are relevant for our test data. Problems starts when SMEs create their own apps which are using real data. The keys used to be different than default teal keys, but the keys follow GDSR requirements. For example ADLB keys areSTUDYID USUBJID PARAMCD BASETYPE AVISITN ATPTN DTYPE ADTM LBSEQ ASPIDvs tealSTUDYID USUBJID PARAMCD AVISIT. SMEs require the change in the keys to fit GDSR standards which complicates the design little bit.If we go back to the last example which is using
ADLB. Extending set of keys will result in joining by keys which have not been filtered-out. This means that ANLs will be joined by"STUDYID", "USUBJID", "ATPTN", "DTYPE", "ADTM", "LBSEQ", "ASPID"if we put onlyPARAMCDandAVISITNas afilter_spec.Changing the keys only will result in:
PARAMCDand/orAVISITfilter_specmight fail because output ANL will be merged on a columns which have a different values across the subjects. One can run above code and see that the result of inner_join is empty data.frame. Also our problem is that GDSR keys are not the same what we think they are. Some of the keys in the datasets does not make any row unique adding them - which means that for us they are just values. For exampleAVISITNis a number of a patient visit whileADTMcan be a data of the visit. This means that addingADTMto the keys does not make any row unique, becauseSTUDYID,USUBJID,PARAMCDandAVISITNmake them unique already.Above is not the case of our sample datasets only, but might occur also in real data see the issue.
drop_keys = TRUEto avoid cartesian join.Because keys/data_merge have been problematic since introduction, we need to simplify the way to handle them before reaching wider users base.
Proposition
Change the keys in
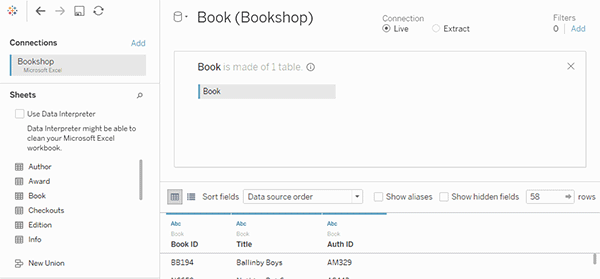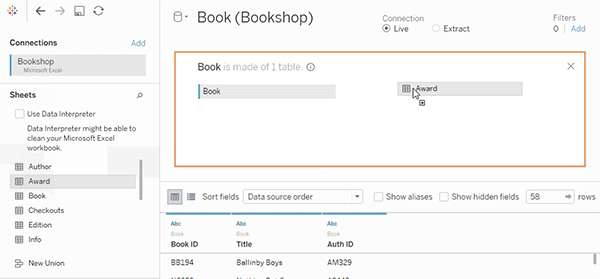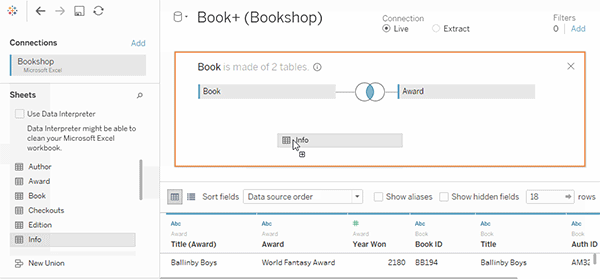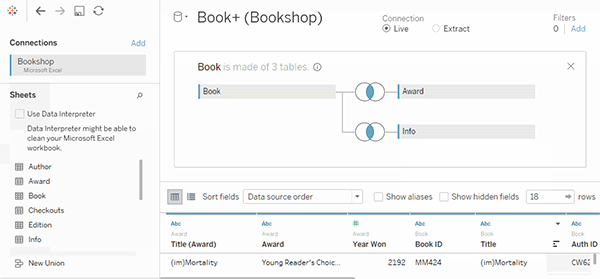
tealaccording to GDSR and implement merge module to visualize the join keys.See a [good example]](https://help.tableau.com/current/pro/desktop/en-us/joining_tables.htm)
Otherwise if we want to keep tea.devel as is we need to fix the documentation to visualize multiple scenarios like:
Current documentation in my opinion is not sufficient.
The text was updated successfully, but these errors were encountered: