See also:
Contents
In this tutorial we’ll be looking at the TAF workflow, which is centered on R scripts that are run sequentially. They structure the stock assessment into separate steps and what we’ll end up with is clean, organized, and reproducible assessments.
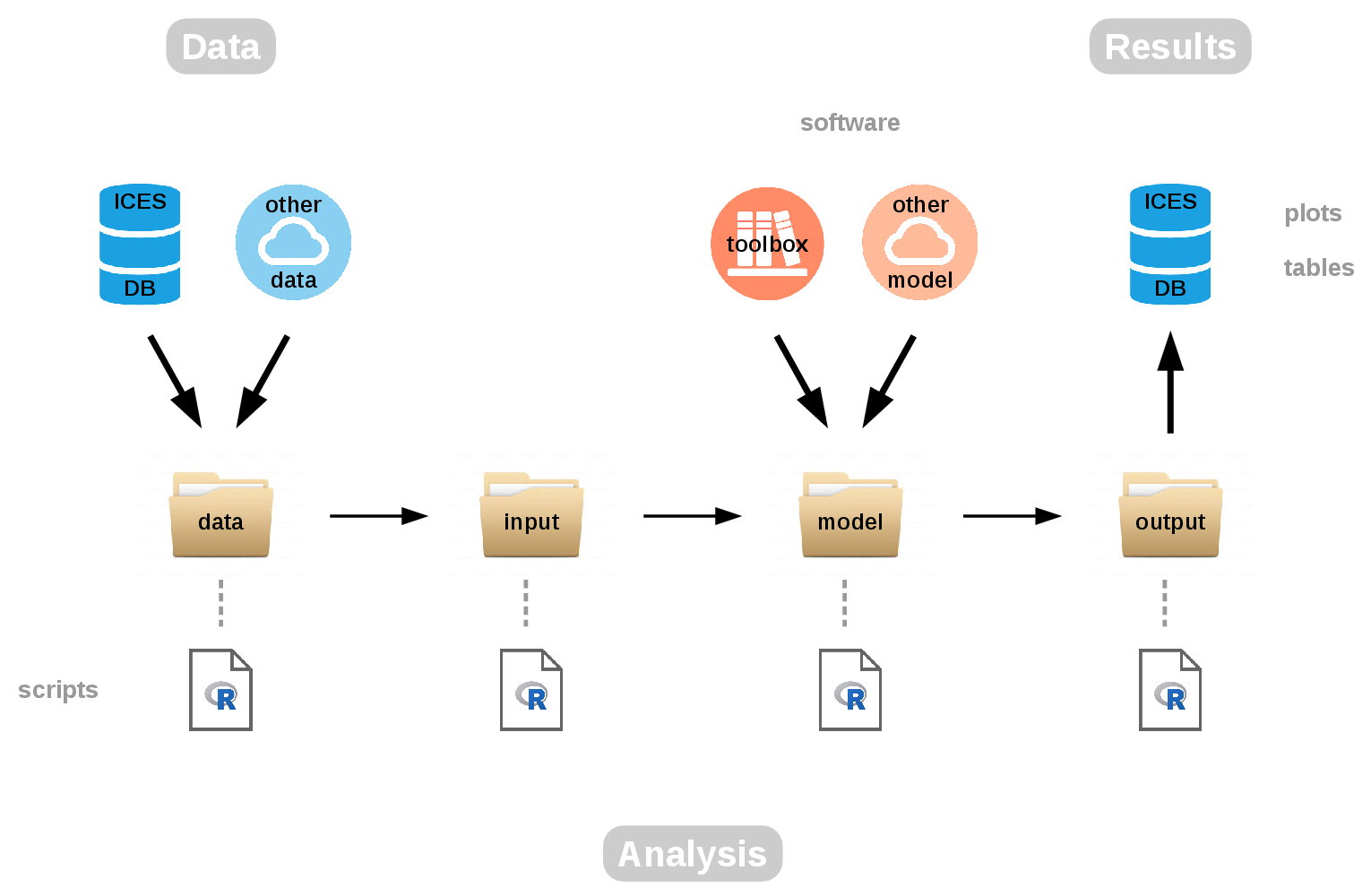
The aim of TAF is to implement a framework to organize data, methods, and results used in ICES assessments, so they’re easy to find and rerun later with new data. If you look at the diagram showing the TAF workflow and the different components, it’s about going from data to analysis and results.
Figure 1: TAF workflow
We start with getting data from ICES databases and other data sources.
The first step in the data folder is to gather the data, and to filter
and preprocess the data that will finally be used in the assessment.
That’s one of the major aims of TAF, to document and script this process
of preparing the data. Describing where they came from and what was done
to them before they were entered in the assessment model.
Moving on to the input folder, the task here is to convert the data
from the most general format, crosstab year by age usually, into the
model-specific format. That will depend on the model: it can be one big
text file, an R list, or a number of input text files, whatever the
model will read. The model folder is about running the model. The
model will be coming from either a toolbox of commonly used models, or
any model can be used within this folder.
The final step, output, is where we convert the model-specific output
into more general text files, things like numbers at age or fishing
mortalities, SSB, and recruitment. These results are then uploaded into
the ICES databases: the stock assessment graphs, tables and so forth.
Behind each of those folders, data, input, model, and output,
there is an R script that governs what takes place. Let’s take a look at
those scripts in more detail.
The first one is data.R. That’s where we preprocess the data and write
out what we call TAF data tables. They’re very simple crosstab text
files, comma separated values. The next step is input.R, where we
convert those data to the model-specific format, writing out the model
input files.
In model.R we run the analysis, often just invoking a shell command or
an R package to run the model, and the results will be written out as
output files. These output files will often contain information about
likelihoods or gradients, and other information we don’t really need. So
we extract the results of interest in output.R, things like numbers at
age and fishing mortalities, and we write them out as text files.
Other scripts that we’ll be working with are report.R, which is an
optional script where scientists can prepare any plots and tables that
they’re going to put in the report, and finally there’s upload.R, a
very short script describing the data that are uploaded into the TAF
system.
What we’re going to do for the rest of the tutorial is to go through the actual analysis behind the 2015 ICES advice for North Sea spotted ray. The R scripts can be found on GitHub at https://github.com/ices-taf/2015_rjm-347d and if you want to work along while you read the tutorial, you can download and work with them on your own computer.
Let’s just dive into data.R. At the top of the script we have comments
reminding us what is the purpose of the script: to preprocess the data
and to write out the TAF data tables. In the comment we also write the
state before the script is run and after the script is run, in terms of
the files and where they are. So we start with catch.csv and
surveys_all.csv in the TAF database. After the script is run we’ll
have catch.csv, summary.csv, and survey.csv, all found in a new
folder called data.
## Preprocess data, write TAF data tables
## Before: catch.csv, surveys_all.csv (TAF database)
## After: catch.csv, summary.csv, survey.csv (data)
library(icesTAF)
mkdir("data")
url <- "https://raw.githubusercontent.com/ices-taf/ftp/master/wgef/2015/rjm-347d/raw/"
## Download data, select years and surveys of interest
catch <- read.taf(paste0(url, "catch.csv"))
survey <- read.taf(paste0(url, "surveys_all.csv"))
survey <- survey[survey$Year %in% 1993:2014, names(survey) != "Unknown"]
## Scale each survey to average 1, combine index as average of three surveys
survey[-1] <- sapply(survey[-1], function(x) x/mean(x, na.rm=TRUE))
survey$Index <- rowMeans(survey[-1])
## Finalize tables
row.names(survey) <- NULL
summary <- data.frame(Year=survey$Year, Catch=NA, Index=survey$Index)
summary$Catch[summary$Year %in% catch$Year] <- catch$Catch
## Write tables to data directory
setwd("data")
write.taf(catch, "catch.csv")
write.taf(survey, "survey.csv")
write.taf(summary, "summary.csv")
setwd("..")Listing 1: data.R
We start by loading the icesTAF package and create an empty directory.
We next download the data, the catch and the survey, and we start
preprocessing the data. We select the years of interest and the surveys
of interest, scale the surveys and create a combined index, as the
average of the three surveys. This combined index will be used as input
data for the assessment. We finalize the tables and we write them out to
the data directory.
What we have done is to create a data folder containing the data that
will be used in the assessment. In catch.csv we have the catch
history, in summary.csv we have combined the catch history with the
index that will be used, and survey.csv documents how the index is
calculated.
## Convert data to model format, write model input files
## Before: catch.csv, survey.csv (data)
## After: input.RData (input)
library(icesTAF)
mkdir("input")
## Get catch and survey data
catch <- read.taf("data/catch.csv")
survey <- read.taf("data/survey.csv")
save(catch, survey, file="input/input.RData")Listing 2: input.R
The next script is input.R. It’s a short script, where we’ll convert
the data to model format and write out the model input files. In other
words, we’ll start with catch and survey in the data folder, but after
the script is run we’ll have input.RData in a folder called input.
As before, we load the icesTAF package and create the directory. We
then simply fetch the catch and the survey data frames, and save them
together in one file, input.RData.
The third script model.R runs the model, and the results will be
written out as dls.txt inside the model folder. Now it’s not enough
just to have the icesTAF package. We also use a package called
icesAdvice, containing the function that we’ll be using to run the
analysis, DLS3.2.
## Run analysis, write model results
## Before: input.RData (input)
## After: dls.txt (model)
library(icesAdvice)
library(icesTAF)
mkdir("model")
## Get data
load("input/input.RData")
## Apply DLS method 3.2
i1 <- survey$Index[nrow(survey)-(6:2)] # five year period from n-6 to n-2
dls <- DLS3.2(mean(catch$Catch), survey$Index, i1=i1)
write.dls(dls, "model/dls.txt")Listing 3: model.R
We start by creating an empty folder, then get the data from the
previous step, apply DLS method 3.2, and the results are found in the
model folder as dls.txt. It outlines the computations behind the
advice. The advice is 291 t, coming from the last advice of 243 t and a
series of survey indices. On average they’ve been going up by 43%, and
the DLS 3.2 rule is that we’re not going to increase the advice by 43%,
but rather by maximum of 20%, so the advice is 291 t.
## Extract results of interest, write TAF output tables
## Before: dls.txt (model)
## After: dls.txt (output)
library(icesTAF)
mkdir("output")
## Copy DLS results to output directory
cp("model/dls.txt", "output")Listing 4: output.R
The output.R script is about extracting those results of interest, and
writing out the TAF output tables. We read in dls.txt and simply copy
it to the output folder. In more complicated stock assessments this
would of course take more steps, but here we just copy between model
and output.
Finally, in report.R, we’re going to prepare plots and tables that
could be included in the stock assessment report. Taking the summary
from the data step, we’ll plot the survey as a PNG file. So we load
the icesTAF package, create an empty directory report, read in the
summary, and create the plot. We also write out the summary table, but
this time rounding the catch and the index values, to make it look
better in a report.
## Prepare plots and tables for report
## Before: summary.csv (data)
## After: summary.csv, survey.png (report)
library(icesTAF)
mkdir("report")
summary <- read.taf("data/summary.csv")
## Plot
taf.png("survey")
plot(summary$Year, summary$Index, type="b", lty=3, ylim=lim(summary$Index),
yaxs="i", main="Survey", xlab="Year", ylab="Index",
panel.first=grid(lwd=2))
dev.off()
## Table
summary <- rnd(summary, "Catch")
summary <- rnd(summary, "Index", 3)
write.taf(summary, "report/summary.csv")Listing 5: report.R
Inside the report folder, we now have summary.csv and survey.png.
The survey plot can be pasted into the report. In the summary table, we
have rounded the indices to three decimals, so it’s also ready for the
report.
In the R scripts we’ve been using some commands from ICES packages.
Let’s take a look at, for example, DLS3.2. The help page describes
it as a function to apply ICES method 3.2, and it has some good
guidelines and references on how to use that function.
library(icesAdvice)
help(DLS3.2)We’ve also been using some R functions from the icesTAF package. If we
take a look at the main help page for the icesTAF package, it lists
all the functions by group. Some of them we’ve been using to read and
write files, and we’ve been using cp and mkdir to manipulate files.
We used taf.png to open a PNG graphics device, to draw an image and
write it in PNG format.
library(icesTAF)
help(icesTAF)Most of the icesTAF functions are very short and simple, but they’re
simply there to make the scripts look more readable. They’re convenient
shorthand functions, to get to the point without some boilerplate code
that’s needed. For example, in report.R we were using taf.png so it
uses the suggested image size and so forth.
We also see in the help page some functions to run the scripts:
sourceTAF to run a single script and sourceAll to run all of them.
Starting from scratch, we can run the scripts one by one:
sourceTAF("data.R")
sourceTAF("input.R")
sourceTAF("model.R")
sourceTAF("output.R")
sourceTAF("report.R")The sourceTAF function is very similar to the base R function
source. It adds a helpful message, showing the time and what it’s
doing. It is often convenient to use sourceAll to run all of the TAF
scripts:
sourceAll()That’s how final assessments will be run on TAF once they’re uploaded,
using sourceAll.
In this tutorial we have learned about the overall TAF workflow. We used as an example the North Sea spotted ray, a fully scripted analysis. We also have on the GitHub TAF page other examples that can be studied in the same way: Icelandic haddock, North Sea cod, and Eastern Channel plaice.
They are all age-based assessments: the Eastern Channel plaice uses the FLR suite of R packages, and the North Sea cod is a SAM model. The Icelandic haddock is an AD Model Builder age-based model, and we’ll be adding more examples here as we go.
In a future tutorial we’ll be covering the TAF web interface, where assessments can be browsed, run, and modified, and how web services can be used to upload, download, and run models.