diff --git a/README.md b/README.md
index 99f2f14..29d45e7 100644
--- a/README.md
+++ b/README.md
@@ -2,55 +2,42 @@
Data-Driven Optimization of Mass Spectrometry Methods
+


* [Website](https://do-ms.slavovlab.net)
* [Get started now](#getting-started)
* [Download](https://github.com/SlavovLab/DO-MS/releases/latest)
-* [Manuscript](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039)
+* [BioArxiv Preprint](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1)
- +
+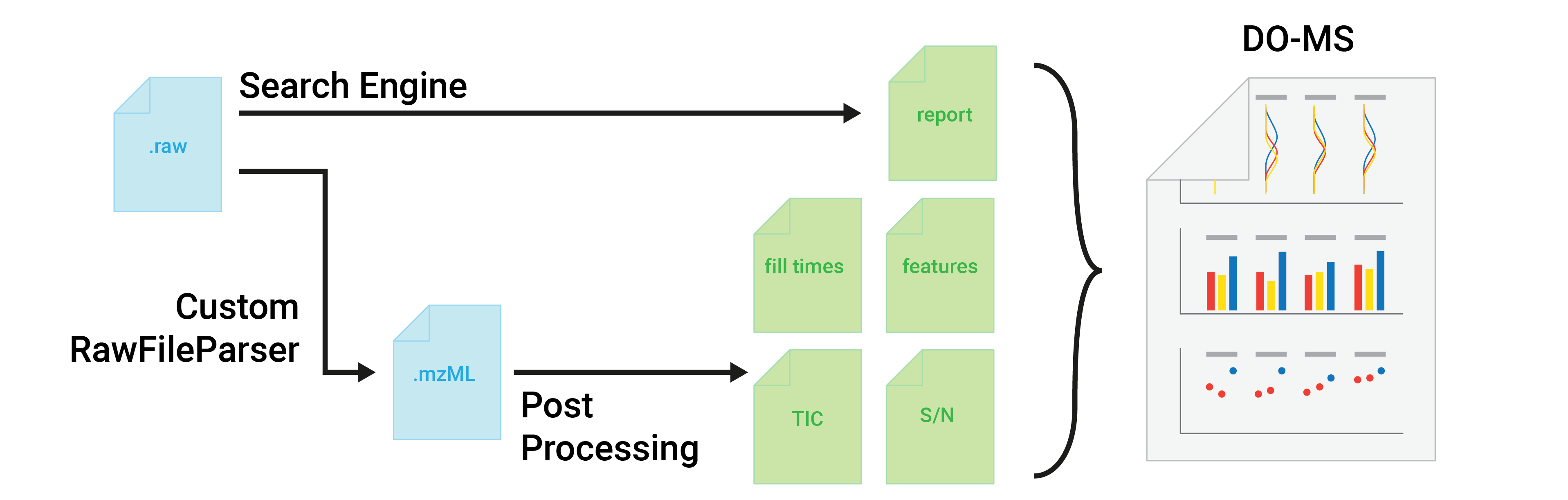 ## Getting Started
Please read our detailed getting started guides:
-* [Getting started on the application](https://do-ms.slavovlab.net/docs/getting-started-application)
-* [Getting started on the command-line](https://do-ms.slavovlab.net/docs/getting-started-command-line)
+* [Getting started with DIA preprocessing](https://do-ms.slavovlab.net/docs/getting-started-preprocessing)
+* [Getting started with DIA reports](https://do-ms.slavovlab.net/docs/getting-started-dia-app)
+* [Getting started with DDA reports](https://do-ms.slavovlab.net/docs/getting-started-dda-app)
### Requirements
-
-This application has been tested on R >= 4.0.0 and R <= 4.0.2, OSX 10.14 / Windows 7/8/10. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and MaxQuant < 2.0
+This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10/11. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and DIA-NN > 1.8.1.
The application suffers from visual glitches when displayed on unsupported older browsers (such as IE9 commonly packaged with RStudio on Windows). Please use IE >= 11, Firefox, or Chrome for the best user experience.
-### Installation
-
-Install this application by downloading it from the [release page](https://github.com/SlavovLab/DO-MS/releases/latest).
-
-### Running
+### Running the Interactive Application
The easiest way to run the app is directly through RStudio, by opening the `DO-MS.Rproj` Rproject file
-
## Getting Started
Please read our detailed getting started guides:
-* [Getting started on the application](https://do-ms.slavovlab.net/docs/getting-started-application)
-* [Getting started on the command-line](https://do-ms.slavovlab.net/docs/getting-started-command-line)
+* [Getting started with DIA preprocessing](https://do-ms.slavovlab.net/docs/getting-started-preprocessing)
+* [Getting started with DIA reports](https://do-ms.slavovlab.net/docs/getting-started-dia-app)
+* [Getting started with DDA reports](https://do-ms.slavovlab.net/docs/getting-started-dda-app)
### Requirements
-
-This application has been tested on R >= 4.0.0 and R <= 4.0.2, OSX 10.14 / Windows 7/8/10. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and MaxQuant < 2.0
+This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10/11. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and DIA-NN > 1.8.1.
The application suffers from visual glitches when displayed on unsupported older browsers (such as IE9 commonly packaged with RStudio on Windows). Please use IE >= 11, Firefox, or Chrome for the best user experience.
-### Installation
-
-Install this application by downloading it from the [release page](https://github.com/SlavovLab/DO-MS/releases/latest).
-
-### Running
+### Running the Interactive Application
The easiest way to run the app is directly through RStudio, by opening the `DO-MS.Rproj` Rproject file
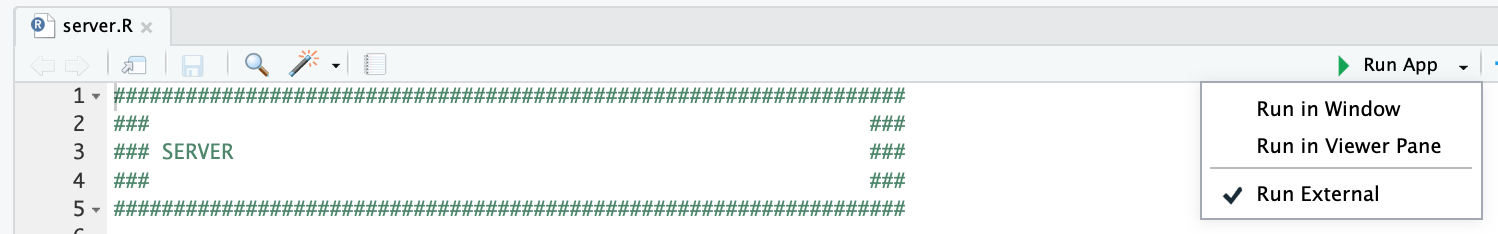
- +{: width="70%" .center-image}
and clicking the "Run App" button at the top of the application, after opening the `server.R` file. We recommend checking the "Run External" option to open the application in your default browser instead of the RStudio Viewer.
-
+{: width="70%" .center-image}
and clicking the "Run App" button at the top of the application, after opening the `server.R` file. We recommend checking the "Run External" option to open the application in your default browser instead of the RStudio Viewer.
- +{: width="70%" .center-image}
You can also start the application by running the `start_server.R` script.
-### Automated Report Generation
-
-You can automatically generate PDF/HTML reports without having to launch the server by running the `do-ms_cmd.R` script, like so:
-
-```
-$ Rscript do-ms_cmd.R config_file.yaml
-```
-
-This requires a configuration file, and you can [find an example one here](https://github.com/SlavovLab/DO-MS/blob/master/example/config_file.yaml). See [Automating Report Generation](https://do-ms.slavovlab.net/docs/automation) for more details and instructions.
-
### Customization
DO-MS is designed to be easily user-customizable for in-house proteomics workflows. Please see [Building Your Own Modules](https://do-ms.slavovlab.net/docs/build-your-own) for more details.
@@ -59,9 +46,9 @@ DO-MS is designed to be easily user-customizable for in-house proteomics workflo
Please see [Hosting as a Server](https://do-ms.slavovlab.net/docs/hosting-as-server) for more details.
-### Search Engines Other Than MaxQuant
+### Supporting other Search Engines
-This application is currently maintained for MaxQuant >= 1.6.0.16. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
+This application is currently maintained for (MaxQuant)[https://www.nature.com/articles/nbt.1511] >= 1.6.0.16 and (DIA-NN)[https://www.nature.com/articles/s41592-019-0638-x] >= 1.8. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
### Can I use this for Metabolomics, Lipidomics, etc... ?
@@ -71,13 +58,8 @@ While the base library of modules are based around bottom-up proteomics by LC-MS
## About the project
-
-
The manuscript for this tool is published at the Journal of Proteome Research: [https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039)
-
-The manuscript is also freely available on bioRxiv: [https://www.biorxiv.org/content/10.1101/512152v1](https://www.biorxiv.org/content/10.1101/512152v1).
+The manuscript for the extended version 2.0 can be found on bioArxiv: [https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1)
Contact the authors by email: [nslavov\{at\}northeastern.edu](mailto:nslavov@northeastern.edu).
@@ -87,21 +69,11 @@ DO-MS is distributed by an [MIT license](https://github.com/SlavovLab/DO-MS/blob
### Contributing
-Please feel free to contribute to this project by opening an issue or pull request.
-
-
-
-
+Please feel free to contribute to this project by opening an issue or pull request in the [GitHub repository](https://github.com/SlavovLab/DO-MS).
-------------
## Help!
-For any bugs, questions, or feature requests,
+For any bugs, questions, or feature requests,
please use the [GitHub issue system](https://github.com/SlavovLab/DO-MS/issues) to contact the developers.
diff --git a/docs/index.md b/docs/index.md
index 68ff4de..d2f82e2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -33,7 +33,8 @@ Install this application by downloading it from the [release page](https://githu
## Getting Started
Please read our detailed getting started guides:
* [Getting started with DIA preprocessing]({{site.baseurl}}/docs/getting-started-preprocessing)
-* [Getting started on the application]({{site.baseurl}}/docs/getting-started-application)
+* [Getting started with DIA reports]({{site.baseurl}}/docs/getting-started-dia-app)
+* [Getting started with DDA reports]({{site.baseurl}}/docs/getting-started-dda-app)
### Requirements
+{: width="70%" .center-image}
You can also start the application by running the `start_server.R` script.
-### Automated Report Generation
-
-You can automatically generate PDF/HTML reports without having to launch the server by running the `do-ms_cmd.R` script, like so:
-
-```
-$ Rscript do-ms_cmd.R config_file.yaml
-```
-
-This requires a configuration file, and you can [find an example one here](https://github.com/SlavovLab/DO-MS/blob/master/example/config_file.yaml). See [Automating Report Generation](https://do-ms.slavovlab.net/docs/automation) for more details and instructions.
-
### Customization
DO-MS is designed to be easily user-customizable for in-house proteomics workflows. Please see [Building Your Own Modules](https://do-ms.slavovlab.net/docs/build-your-own) for more details.
@@ -59,9 +46,9 @@ DO-MS is designed to be easily user-customizable for in-house proteomics workflo
Please see [Hosting as a Server](https://do-ms.slavovlab.net/docs/hosting-as-server) for more details.
-### Search Engines Other Than MaxQuant
+### Supporting other Search Engines
-This application is currently maintained for MaxQuant >= 1.6.0.16. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
+This application is currently maintained for (MaxQuant)[https://www.nature.com/articles/nbt.1511] >= 1.6.0.16 and (DIA-NN)[https://www.nature.com/articles/s41592-019-0638-x] >= 1.8. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
### Can I use this for Metabolomics, Lipidomics, etc... ?
@@ -71,13 +58,8 @@ While the base library of modules are based around bottom-up proteomics by LC-MS
## About the project
-
-
The manuscript for this tool is published at the Journal of Proteome Research: [https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039)
-
-The manuscript is also freely available on bioRxiv: [https://www.biorxiv.org/content/10.1101/512152v1](https://www.biorxiv.org/content/10.1101/512152v1).
+The manuscript for the extended version 2.0 can be found on bioArxiv: [https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1)
Contact the authors by email: [nslavov\{at\}northeastern.edu](mailto:nslavov@northeastern.edu).
@@ -87,21 +69,11 @@ DO-MS is distributed by an [MIT license](https://github.com/SlavovLab/DO-MS/blob
### Contributing
-Please feel free to contribute to this project by opening an issue or pull request.
-
-
-
-
+Please feel free to contribute to this project by opening an issue or pull request in the [GitHub repository](https://github.com/SlavovLab/DO-MS).
-------------
## Help!
-For any bugs, questions, or feature requests,
+For any bugs, questions, or feature requests,
please use the [GitHub issue system](https://github.com/SlavovLab/DO-MS/issues) to contact the developers.
diff --git a/docs/index.md b/docs/index.md
index 68ff4de..d2f82e2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -33,7 +33,8 @@ Install this application by downloading it from the [release page](https://githu
## Getting Started
Please read our detailed getting started guides:
* [Getting started with DIA preprocessing]({{site.baseurl}}/docs/getting-started-preprocessing)
-* [Getting started on the application]({{site.baseurl}}/docs/getting-started-application)
+* [Getting started with DIA reports]({{site.baseurl}}/docs/getting-started-dia-app)
+* [Getting started with DDA reports]({{site.baseurl}}/docs/getting-started-dda-app)
### Requirements
 +
+ ## Getting Started
Please read our detailed getting started guides:
-* [Getting started on the application](https://do-ms.slavovlab.net/docs/getting-started-application)
-* [Getting started on the command-line](https://do-ms.slavovlab.net/docs/getting-started-command-line)
+* [Getting started with DIA preprocessing](https://do-ms.slavovlab.net/docs/getting-started-preprocessing)
+* [Getting started with DIA reports](https://do-ms.slavovlab.net/docs/getting-started-dia-app)
+* [Getting started with DDA reports](https://do-ms.slavovlab.net/docs/getting-started-dda-app)
### Requirements
-
-This application has been tested on R >= 4.0.0 and R <= 4.0.2, OSX 10.14 / Windows 7/8/10. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and MaxQuant < 2.0
+This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10/11. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and DIA-NN > 1.8.1.
The application suffers from visual glitches when displayed on unsupported older browsers (such as IE9 commonly packaged with RStudio on Windows). Please use IE >= 11, Firefox, or Chrome for the best user experience.
-### Installation
-
-Install this application by downloading it from the [release page](https://github.com/SlavovLab/DO-MS/releases/latest).
-
-### Running
+### Running the Interactive Application
The easiest way to run the app is directly through RStudio, by opening the `DO-MS.Rproj` Rproject file
-
## Getting Started
Please read our detailed getting started guides:
-* [Getting started on the application](https://do-ms.slavovlab.net/docs/getting-started-application)
-* [Getting started on the command-line](https://do-ms.slavovlab.net/docs/getting-started-command-line)
+* [Getting started with DIA preprocessing](https://do-ms.slavovlab.net/docs/getting-started-preprocessing)
+* [Getting started with DIA reports](https://do-ms.slavovlab.net/docs/getting-started-dia-app)
+* [Getting started with DDA reports](https://do-ms.slavovlab.net/docs/getting-started-dda-app)
### Requirements
-
-This application has been tested on R >= 4.0.0 and R <= 4.0.2, OSX 10.14 / Windows 7/8/10. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and MaxQuant < 2.0
+This application has been tested on R >= 3.5.0, OSX 10.14 / Windows 7/8/10/11. R can be downloaded from the main [R Project page](https://www.r-project.org/) or downloaded with the [RStudio Application](https://www.rstudio.com/products/rstudio/download/). All modules are maintained for MaxQuant >= 1.6.0.16 and DIA-NN > 1.8.1.
The application suffers from visual glitches when displayed on unsupported older browsers (such as IE9 commonly packaged with RStudio on Windows). Please use IE >= 11, Firefox, or Chrome for the best user experience.
-### Installation
-
-Install this application by downloading it from the [release page](https://github.com/SlavovLab/DO-MS/releases/latest).
-
-### Running
+### Running the Interactive Application
The easiest way to run the app is directly through RStudio, by opening the `DO-MS.Rproj` Rproject file
- +{: width="70%" .center-image}
and clicking the "Run App" button at the top of the application, after opening the `server.R` file. We recommend checking the "Run External" option to open the application in your default browser instead of the RStudio Viewer.
-
+{: width="70%" .center-image}
and clicking the "Run App" button at the top of the application, after opening the `server.R` file. We recommend checking the "Run External" option to open the application in your default browser instead of the RStudio Viewer.
- +{: width="70%" .center-image}
You can also start the application by running the `start_server.R` script.
-### Automated Report Generation
-
-You can automatically generate PDF/HTML reports without having to launch the server by running the `do-ms_cmd.R` script, like so:
-
-```
-$ Rscript do-ms_cmd.R config_file.yaml
-```
-
-This requires a configuration file, and you can [find an example one here](https://github.com/SlavovLab/DO-MS/blob/master/example/config_file.yaml). See [Automating Report Generation](https://do-ms.slavovlab.net/docs/automation) for more details and instructions.
-
### Customization
DO-MS is designed to be easily user-customizable for in-house proteomics workflows. Please see [Building Your Own Modules](https://do-ms.slavovlab.net/docs/build-your-own) for more details.
@@ -59,9 +46,9 @@ DO-MS is designed to be easily user-customizable for in-house proteomics workflo
Please see [Hosting as a Server](https://do-ms.slavovlab.net/docs/hosting-as-server) for more details.
-### Search Engines Other Than MaxQuant
+### Supporting other Search Engines
-This application is currently maintained for MaxQuant >= 1.6.0.16. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
+This application is currently maintained for (MaxQuant)[https://www.nature.com/articles/nbt.1511] >= 1.6.0.16 and (DIA-NN)[https://www.nature.com/articles/s41592-019-0638-x] >= 1.8. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
### Can I use this for Metabolomics, Lipidomics, etc... ?
@@ -71,13 +58,8 @@ While the base library of modules are based around bottom-up proteomics by LC-MS
## About the project
-
-
The manuscript for this tool is published at the Journal of Proteome Research: [https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039)
-
-The manuscript is also freely available on bioRxiv: [https://www.biorxiv.org/content/10.1101/512152v1](https://www.biorxiv.org/content/10.1101/512152v1).
+The manuscript for the extended version 2.0 can be found on bioArxiv: [https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1)
Contact the authors by email: [nslavov\{at\}northeastern.edu](mailto:nslavov@northeastern.edu).
@@ -87,21 +69,11 @@ DO-MS is distributed by an [MIT license](https://github.com/SlavovLab/DO-MS/blob
### Contributing
-Please feel free to contribute to this project by opening an issue or pull request.
-
-
-
-
+Please feel free to contribute to this project by opening an issue or pull request in the [GitHub repository](https://github.com/SlavovLab/DO-MS).
-------------
## Help!
-For any bugs, questions, or feature requests,
+For any bugs, questions, or feature requests,
please use the [GitHub issue system](https://github.com/SlavovLab/DO-MS/issues) to contact the developers.
diff --git a/docs/index.md b/docs/index.md
index 68ff4de..d2f82e2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -33,7 +33,8 @@ Install this application by downloading it from the [release page](https://githu
## Getting Started
Please read our detailed getting started guides:
* [Getting started with DIA preprocessing]({{site.baseurl}}/docs/getting-started-preprocessing)
-* [Getting started on the application]({{site.baseurl}}/docs/getting-started-application)
+* [Getting started with DIA reports]({{site.baseurl}}/docs/getting-started-dia-app)
+* [Getting started with DDA reports]({{site.baseurl}}/docs/getting-started-dda-app)
### Requirements
+{: width="70%" .center-image}
You can also start the application by running the `start_server.R` script.
-### Automated Report Generation
-
-You can automatically generate PDF/HTML reports without having to launch the server by running the `do-ms_cmd.R` script, like so:
-
-```
-$ Rscript do-ms_cmd.R config_file.yaml
-```
-
-This requires a configuration file, and you can [find an example one here](https://github.com/SlavovLab/DO-MS/blob/master/example/config_file.yaml). See [Automating Report Generation](https://do-ms.slavovlab.net/docs/automation) for more details and instructions.
-
### Customization
DO-MS is designed to be easily user-customizable for in-house proteomics workflows. Please see [Building Your Own Modules](https://do-ms.slavovlab.net/docs/build-your-own) for more details.
@@ -59,9 +46,9 @@ DO-MS is designed to be easily user-customizable for in-house proteomics workflo
Please see [Hosting as a Server](https://do-ms.slavovlab.net/docs/hosting-as-server) for more details.
-### Search Engines Other Than MaxQuant
+### Supporting other Search Engines
-This application is currently maintained for MaxQuant >= 1.6.0.16. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
+This application is currently maintained for (MaxQuant)[https://www.nature.com/articles/nbt.1511] >= 1.6.0.16 and (DIA-NN)[https://www.nature.com/articles/s41592-019-0638-x] >= 1.8. Adapting to other search engines is possible but not provided out-of-the-box. Please see [Integrating Other Search Engines ](https://do-ms.slavovlab.net/docs/other-search-engines) for more details.
### Can I use this for Metabolomics, Lipidomics, etc... ?
@@ -71,13 +58,8 @@ While the base library of modules are based around bottom-up proteomics by LC-MS
## About the project
-
-
The manuscript for this tool is published at the Journal of Proteome Research: [https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039](https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00039)
-
-The manuscript is also freely available on bioRxiv: [https://www.biorxiv.org/content/10.1101/512152v1](https://www.biorxiv.org/content/10.1101/512152v1).
+The manuscript for the extended version 2.0 can be found on bioArxiv: [https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1](https://www.biorxiv.org/content/10.1101/2023.02.02.526809v1)
Contact the authors by email: [nslavov\{at\}northeastern.edu](mailto:nslavov@northeastern.edu).
@@ -87,21 +69,11 @@ DO-MS is distributed by an [MIT license](https://github.com/SlavovLab/DO-MS/blob
### Contributing
-Please feel free to contribute to this project by opening an issue or pull request.
-
-
-
-
+Please feel free to contribute to this project by opening an issue or pull request in the [GitHub repository](https://github.com/SlavovLab/DO-MS).
-------------
## Help!
-For any bugs, questions, or feature requests,
+For any bugs, questions, or feature requests,
please use the [GitHub issue system](https://github.com/SlavovLab/DO-MS/issues) to contact the developers.
diff --git a/docs/index.md b/docs/index.md
index 68ff4de..d2f82e2 100644
--- a/docs/index.md
+++ b/docs/index.md
@@ -33,7 +33,8 @@ Install this application by downloading it from the [release page](https://githu
## Getting Started
Please read our detailed getting started guides:
* [Getting started with DIA preprocessing]({{site.baseurl}}/docs/getting-started-preprocessing)
-* [Getting started on the application]({{site.baseurl}}/docs/getting-started-application)
+* [Getting started with DIA reports]({{site.baseurl}}/docs/getting-started-dia-app)
+* [Getting started with DDA reports]({{site.baseurl}}/docs/getting-started-dda-app)
### Requirements