CellSignal - Disentangling biological signal from experimental noise in cellular images
Full disclosure: Some text in this proposal will be taken word for word from the competition's websites.
Recursion Pharmaceuticals, creators of the industry’s largest dataset of biological images, generated entirely in-house, believes AI has the potential to dramatically improve and expedite the drug discovery process. More specifically, machine learning could help understand how drugs interact with human cells.
This competition is designed to disentangle experimental noise from real biological signals. The goal is to classify images of cells under one of 1,108 different genetic perturbations, and thus eliminate the noise introduced by technical execution and environmental variation between [drug] experiments.
This is a multiclass classification challenge applied to a healthcare related topic. Other papers have been published in the past in this broad field:
- https://arxiv.org/abs/1903.10035
- https://onlinelibrary.wiley.com/doi/abs/10.1002/mrm.27229
- https://pdfs.semanticscholar.org/2583/2df680518e0b36734f54fb640668fe834be8.pdf
- https://onlinelibrary.wiley.com/doi/full/10.1002/mrm.26841
This is a multiclass classification problem.
The inputs will be 512x512 pixels images and the output is a genetic perturbation (siRNA) represented by an integer ranging from 1 to 1,108.
below is an output example
922
The data is available on the Kaggle's competition site https://www.kaggle.com/c/recursion-cellular-image-classification/data . For more information about the dataset, see the competition's website.
The input images are all 512x512 pixels black and white.
identify example_input.png
# --> example_input.png PNG 512x512 512x512+0+0 8-bit Gray 256c 58265B 0.000u 0:00.014Along with images, three other features are provided:
experiment: the cell type and batch numberplate: plate number within the experimentwell: location on the plate
The outcome variable is siRNA, which is a multiclass variable which can be an integer ranging from 1 to 1,108, each integer representing a different genetic disruption.
The data is already splitted into training and test sets. Further splitting will be considered if needed.
One of the main challenges for applying AI to biological microscopy data is that even the most careful replicates of a process will not look identical. This dataset challenges you to develop a model for identifying replicates that is robust to experimental noise.
The same siRNAs (effectively genetic perturbations) have been applied repeatedly to multiple cell lines, for a total of 51 experimental batches. Each batch has four plates, each of which has 308 filled wells. For each well, microscope images were taken at two sites and across six imaging channels. Not every batch will necessarily have every well filled or every siRNA present.
The solution for this problem will likely be resolved with the type of model architecture used in computer vision and image classification, e.g convolutional neural networks. This is a multiclass classification problem, but algorithms and model architectures we have seen in the dogs classification project https://github.com/MichelML/udacity-dog-project/, such as VGG-16 and ResNet-50, will be considered for this project.
As discussed previously, our solution will most likely use a convolutional neural network architecture. We will thus use a vanilla CNN model as our benchmark, such as the one we built during step 3 of the dogs classification project https://github.com/MichelML/udacity-dog-project/blob/master/dog_app.ipynb .
To justify our decision to go along with a CNN architecture, it is to be said ResNet-50 have achieved very good results in multiclass image classification problems in the recent past (see source), having reached 98.87% accuracy when classifying histopathology images.
Submissions will be evaluated on Multiclass Accuracy, which is simply the average number of observations with the correct label.
It is okay to stick with accuracy in our context, as each class has a roughly equal number of data points.
For each id_code in the test set, we will predict the correct siRNA. As per the competition's indications, The file should contain a header and have the following format:
id_code,sirna
HEPG2-08_1_B03,911
HEPG2-08_1_B04,911
etc.
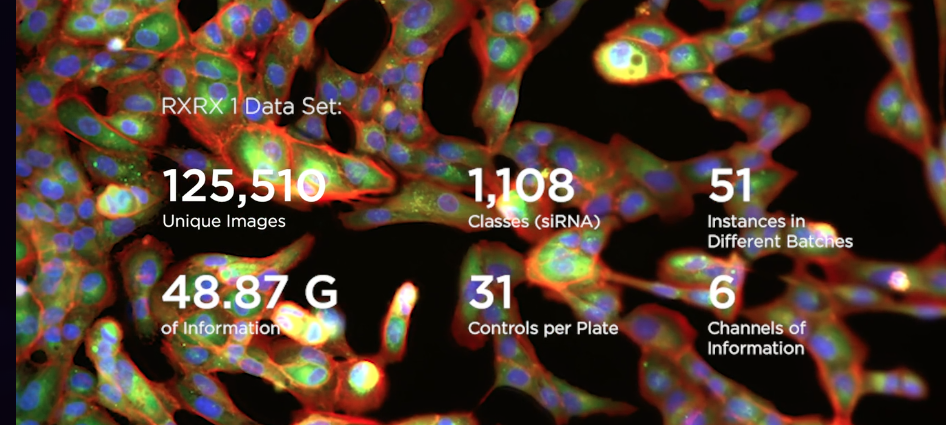
The dataset includes data from 51 instances of the same experiment design executed in different experimental batches. In this experiment, there is 1,108 different siRNAs to knockdown 1,108 different genes.
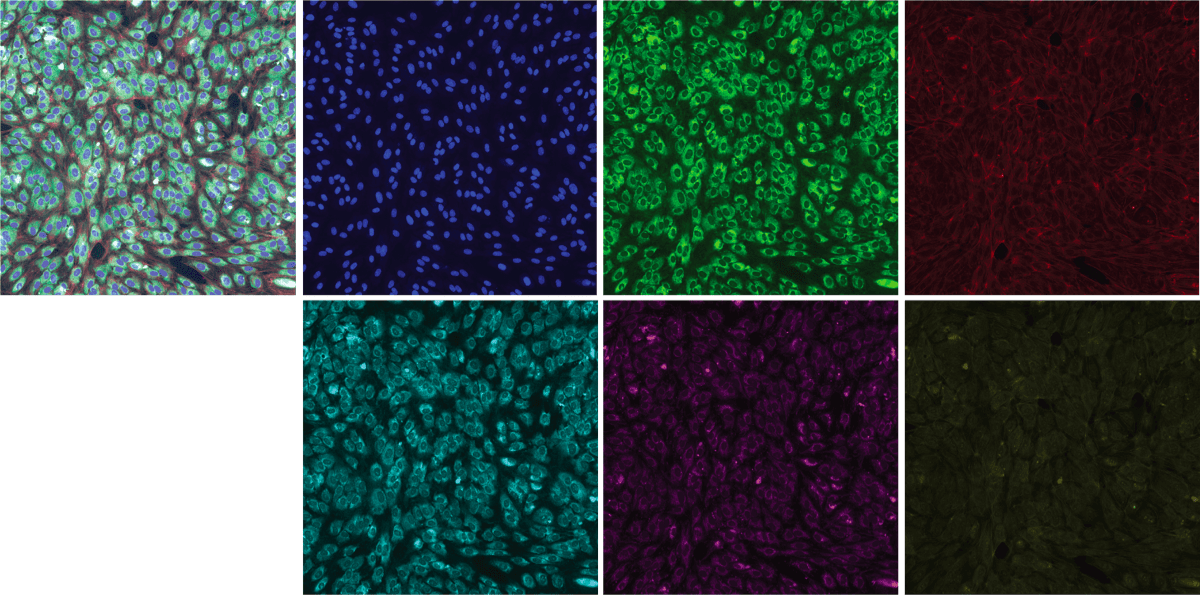
The experiment uses 384-well plates to isolate populations of cells into wells where exactly one of 1,108 different siRNAs is introduced into the well to create distinct genetic conditions. A well is like a single test tube at a small scale, 3.3 mm2. The outer rows and columns of the plate are not used because they are subject to greater environmental effects; so there are 308 used wells on each plate. Thus the experiment consists of 4 total plates. Each plate holds the same 30 control siRNA conditions, 277 different non-control siRNA, and one untreated well. The location of each of the 1,108 non-control siRNA conditions is randomized in each experiment to prevent confounding effects of the location of a particular well (see Plate Effects). Each well in each plate contains two 512 x 512 x 6 images. The images were acquired from two non-overlapping regions of each well. Each of the 6 channels can be assigned a consistent color and composited for ease of reviewing, however the RxRx1 contains the 6-channel images and not the composite images.
- Data cleaning
- Prepare and pre-process the data.
- Explore many different models and short-list the best ones based on training results.
- Fine-tune your models and combine them into a great solution.
- Iterate & final conclusion.
- Remove all images of empty wells (if present), which should be all the images representing the outer rows and columns of plates
- Combine all 6 images representing a given snapshot of each genetic perturbation (target variable, siRNA). There are two snapshot of a genetic perturbation per well. Here is an example of a recombination of a snapshot using coloring and image composition (see Bray & al.):
-
Use progressive resizing to speed up the learning process.
-
Transform categorical variables 1) experiment, 2) plate and 3) well in dummy variables.
-
Since there are two snapshots (site) per well, an extra categorical (dummy) variable could be added for this too.
Since we have images and categorical data, we will want to use both in our multiclass classification model:
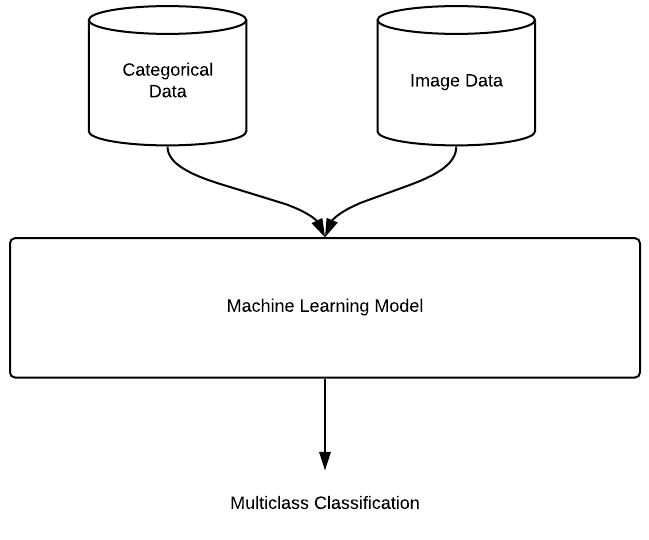
The dataset is comparable to datasets such as ImageNet (ILSVRC2012) which is approximately 155 GB and 1.2m images with 1000 classes. We will try using transfer learning and use the ResNet-50 trained on ImageNet preexisting weights to train our model. The model will start with the pre-trained ResNet-50 model as a fixed feature extractor, where the last convolutional output of ResNet-50 will be fed as input to our additional layers. We will only add a global average pooling layer and a fully connected layer, where the latter will contain one node for each siRNA category (1,108), and will use softmax activation.
Other models known for image classification can be tried at this stage: VGG-19, Inception, Xception, etc.
- Try to improve results tweaking hyperparameters, more epochs, feature engineering, etc.
- Repeat previous steps until no further improvement is made
- Present final conclusions