 ](https://bioinformatics.ccr.cancer.gov/ccbr/)
+[
](https://bioinformatics.ccr.cancer.gov/ccbr/)
+[ ](https://bioinformatics.ccr.cancer.gov/ccbr/)
+## Table of Contents
+
+ - [NEW Releases](#new-releases)
+ - [TOP contributors](#top-contributors)
+ - [About Us](#about-us)
+ - [Our model](#our-model)
+ - [Pipelines](#pipelines)
+ - [Tools](#tools)
+ - [Release History](#release-history)
+ - [Latest Releases of pipelines/tools:](#latest-releases-of-pipelines/tools:)
+ - [Citation](#citation)
+
+## NEW Releases
+
+| Repo Name | Release Name | Release Date | Open Issues |
+|:-----------------------------------------------------|:---------------------------------------------------------------------------|:---------------|--------------:|
+| [spacesavers2](https://github.com/CCBR/spacesavers2) | [v0.14.0](https://github.com/CCBR/spacesavers2/releases/tag/v0.14.0) | 2024-07-16 | 5 |
+| [XAVIER](https://github.com/CCBR/XAVIER) | [v3.0.3](https://github.com/CCBR/XAVIER/releases/tag/v3.0.3) | 2024-07-11 | 9 |
+| [ESCAPE](https://github.com/CCBR/ESCAPE) | [v1.1.2](https://github.com/CCBR/ESCAPE/releases/tag/v1.1.2) | 2024-06-27 | 1 |
+| [permfix](https://github.com/CCBR/permfix) | [v0.6.4](https://github.com/CCBR/permfix/releases/tag/v0.6.4) | 2024-05-07 | 0 |
+| [journal-club](https://github.com/CCBR/journal-club) | [jchelper 0.1.0](https://github.com/CCBR/journal-club/releases/tag/v0.1.0) | 2024-05-07 | 2 |
+| [reports](https://github.com/CCBR/reports) | [ccbr.reports 0.2.0](https://github.com/CCBR/reports/releases/tag/v0.2.0) | 2024-04-30 | 11 |
+
+## TOP contributors
+
+| User | Total Commits | Commits in Last Month | Commits in Last 6 Months |
+|:---------------|----------------:|------------------------:|---------------------------:|
+| kopardev | 4114 | 31 | 528 |
+| kelly-sovacool | 3310 | 122 | 1051 |
+| slsevilla | 1700 | 0 | 119 |
+| skchronicles | 1074 | 0 | 0 |
+| dnousome | 647 | 13 | 137 |
+| kcgfarb | 478 | 28 | 169 |
+| finneyr | 342 | 1 | 20 |
+| samarth8392 | 339 | 20 | 99 |
+| jlac | 307 | 0 | 0 |
+| kvaldez | 222 | 0 | 0 |
+
+## About Us
-### About Us
- 👋 Hi, we're the [**@CCBR**](https://bioinformatics.ccr.cancer.gov/ccbr/), a group of bioinformatics analysts and engineers
- 📖 We build flexible, reproducible, workflows for next-generation sequencing data
- :bulb: We [collaborate](https://abcs-amp.nih.gov/project/request/CCBR/) with [CCR](https://ccr.cancer.gov/) PIs
- 📫 You can reach us at [ccbr_pipeliner@mail.nih.gov](mailto:ccbr_pipeliner@mail.nih.gov)
- 🏁 Check out our [release history](#release-history)
- :link: Our [Zenodo](https://zenodo.org/communities/ccbr) community
+
](https://bioinformatics.ccr.cancer.gov/ccbr/)
+## Table of Contents
+
+ - [NEW Releases](#new-releases)
+ - [TOP contributors](#top-contributors)
+ - [About Us](#about-us)
+ - [Our model](#our-model)
+ - [Pipelines](#pipelines)
+ - [Tools](#tools)
+ - [Release History](#release-history)
+ - [Latest Releases of pipelines/tools:](#latest-releases-of-pipelines/tools:)
+ - [Citation](#citation)
+
+## NEW Releases
+
+| Repo Name | Release Name | Release Date | Open Issues |
+|:-----------------------------------------------------|:---------------------------------------------------------------------------|:---------------|--------------:|
+| [spacesavers2](https://github.com/CCBR/spacesavers2) | [v0.14.0](https://github.com/CCBR/spacesavers2/releases/tag/v0.14.0) | 2024-07-16 | 5 |
+| [XAVIER](https://github.com/CCBR/XAVIER) | [v3.0.3](https://github.com/CCBR/XAVIER/releases/tag/v3.0.3) | 2024-07-11 | 9 |
+| [ESCAPE](https://github.com/CCBR/ESCAPE) | [v1.1.2](https://github.com/CCBR/ESCAPE/releases/tag/v1.1.2) | 2024-06-27 | 1 |
+| [permfix](https://github.com/CCBR/permfix) | [v0.6.4](https://github.com/CCBR/permfix/releases/tag/v0.6.4) | 2024-05-07 | 0 |
+| [journal-club](https://github.com/CCBR/journal-club) | [jchelper 0.1.0](https://github.com/CCBR/journal-club/releases/tag/v0.1.0) | 2024-05-07 | 2 |
+| [reports](https://github.com/CCBR/reports) | [ccbr.reports 0.2.0](https://github.com/CCBR/reports/releases/tag/v0.2.0) | 2024-04-30 | 11 |
+
+## TOP contributors
+
+| User | Total Commits | Commits in Last Month | Commits in Last 6 Months |
+|:---------------|----------------:|------------------------:|---------------------------:|
+| kopardev | 4114 | 31 | 528 |
+| kelly-sovacool | 3310 | 122 | 1051 |
+| slsevilla | 1700 | 0 | 119 |
+| skchronicles | 1074 | 0 | 0 |
+| dnousome | 647 | 13 | 137 |
+| kcgfarb | 478 | 28 | 169 |
+| finneyr | 342 | 1 | 20 |
+| samarth8392 | 339 | 20 | 99 |
+| jlac | 307 | 0 | 0 |
+| kvaldez | 222 | 0 | 0 |
+
+## About Us
-### About Us
- 👋 Hi, we're the [**@CCBR**](https://bioinformatics.ccr.cancer.gov/ccbr/), a group of bioinformatics analysts and engineers
- 📖 We build flexible, reproducible, workflows for next-generation sequencing data
- :bulb: We [collaborate](https://abcs-amp.nih.gov/project/request/CCBR/) with [CCR](https://ccr.cancer.gov/) PIs
- 📫 You can reach us at [ccbr_pipeliner@mail.nih.gov](mailto:ccbr_pipeliner@mail.nih.gov)
- 🏁 Check out our [release history](#release-history)
- :link: Our [Zenodo](https://zenodo.org/communities/ccbr) community
++ Back to Top +
-### Our model +## Our model [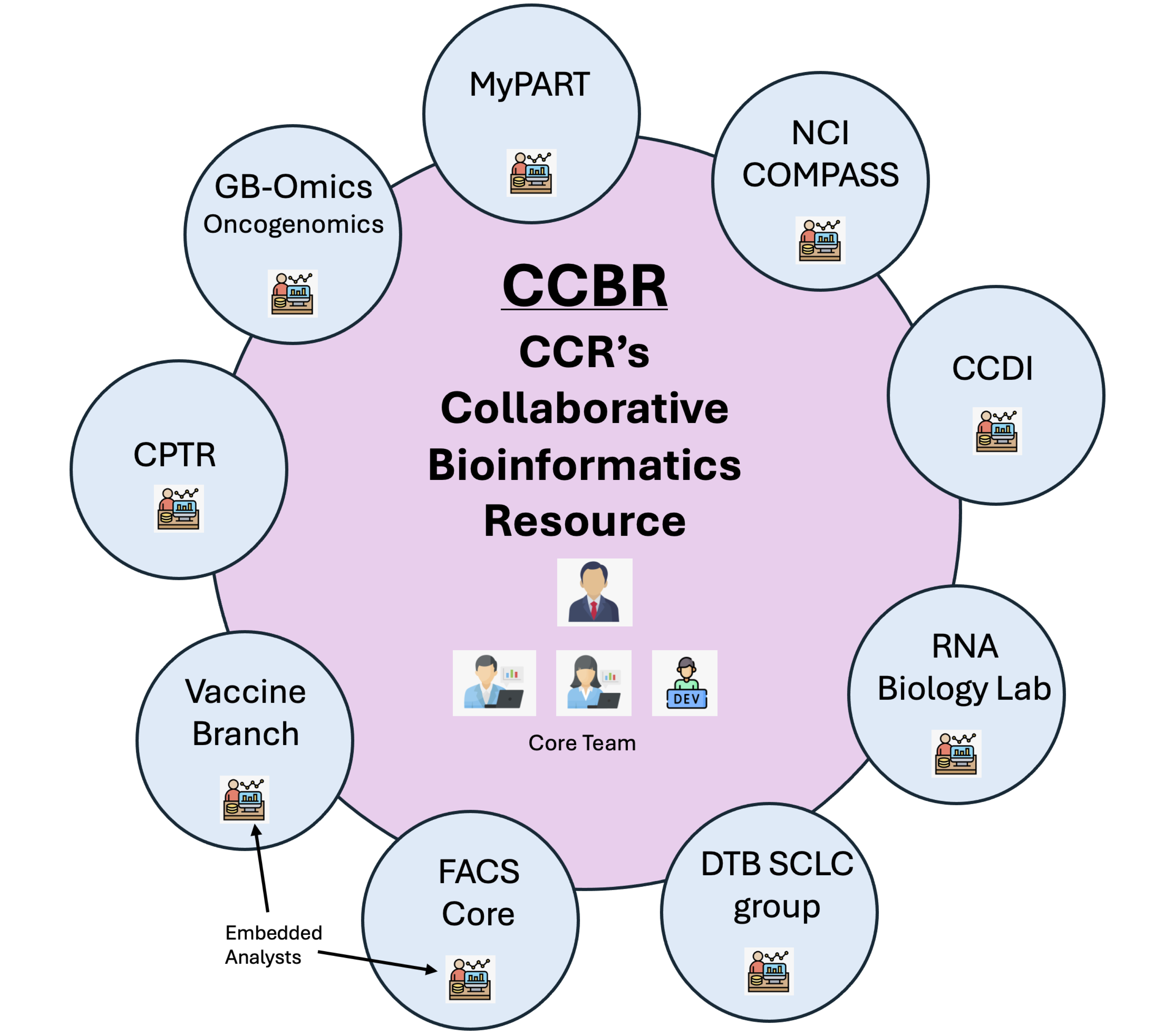 ](https://bioinformatics.ccr.cancer.gov/ccbr/)
-### Pipelines
+
](https://bioinformatics.ccr.cancer.gov/ccbr/)
-### Pipelines
++ Back to Top +
+ +## Pipelines CCBR offers end-to-end analysis pipelines for NGS data analysis. @@ -28,11 +75,13 @@ Here is a list of our prominent pipelines and their release schedule on BIOWULF: | WESSeq2 | [XAVIER](https://github.com/CCBR/XAVIER) | July 21th 2023 | Sep 1st 2023 | | ATACSeq3 | [ASPEN](https://github.com/CCBR/ASPEN) | November 30th 2023 | TBD | | ChIPSeq4 | [CHAMPAGNE](https://github.com/CCBR/CHAMPAGNE) | October 15th 2023 | TBD | -| CRISPRSeq5 | [CRUISE](https://github.com/CCBR/CRUISE) | October 31st 2023 | TBD | +| CRISPRSeq5 | [CRISPIN](https://github.com/CCBR/CRISPIN) | September 31st 2023 | TBD | | CUT&RunSeq6 | [CARLISLE](https://github.com/CCBR/CARLISLE) | October 31st 2023 | TBD | -| circRNASeq7 | [CHARLIE](https://github.com/CCBR/CHARLIE) | March 31st 2024 | TBD | -| scRNASeq8 | [SINCLAIR](https://github.com/CCBR/SINCLAIR) | April 30th 2024 | TBD | -| WGSSeq9 | [LOGAN](https://github.com/CCBR/LOGAN) | May 31st 2024 | TBD | +| EV-Seq10 | [ESCAPE](https://github.com/CCBR/ESCAPE) | March 26th, 2024 | TBD | +| circRNASeq7 | [CHARLIE](https://github.com/CCBR/CHARLIE) | _Jul 31st 2024_ | TBD | +| scRNASeq8 | [SINCLAIR](https://github.com/CCBR/SINCLAIR) | _Sep 30th 2024_ | TBD | +| WGSSeq9 | [LOGAN](https://github.com/CCBR/LOGAN) | _Sep 30th 2024_ | TBD | +| spatialSeq11 | [SPENCER](https://github.com/CCBR/SPENCER) | TBD | TBD | * CLI = Command Line Interface * GUI = Graphical User Interface @@ -53,16 +102,20 @@ Here is a list of our prominent pipelines and their release schedule on BIOWULF: **8** SINCLAIR=_SINgle CelL AnalysIs Resource_ addresses various single cell modalities... eg. single-cell expression, CITESeq, TCR-Seq, etc. - **9** WGSSeq pipeline will soon be CCBR's newest offering. + **9** LOGAN=_whoLe genOme-sequencinG Analysis pipeliNe_ will soon be CCBR's newest offering. + + **10** ESCAPE=_Extracellular veSiCles rnAseq PipelinE_. + + **11** SPENCER=_SPatial seqeENCing Resource_. For any other datatype or pipeline, please [email :mailbox:](mailto:ccbr_pipeliner@mail.nih.gov) us directly to get the conversation started!+ Back to Top +
+## Latest Releases of pipelines/tools: + +| Repo Name | Release Name | Release Date | Open Issues | +|:-------------------------------------------------------------------------------------------------------------------------|:----------------------------------------------------------------------------------------------------------------|:---------------|--------------:| +| [spacesavers2](https://github.com/CCBR/spacesavers2) | [v0.14.0](https://github.com/CCBR/spacesavers2/releases/tag/v0.14.0) | 2024-07-16 | 5 | +| [XAVIER](https://github.com/CCBR/XAVIER) | [v3.0.3](https://github.com/CCBR/XAVIER/releases/tag/v3.0.3) | 2024-07-11 | 9 | +| [ESCAPE](https://github.com/CCBR/ESCAPE) | [v1.1.2](https://github.com/CCBR/ESCAPE/releases/tag/v1.1.2) | 2024-06-27 | 1 | +| [permfix](https://github.com/CCBR/permfix) | [v0.6.4](https://github.com/CCBR/permfix/releases/tag/v0.6.4) | 2024-05-07 | 0 | +| [journal-club](https://github.com/CCBR/journal-club) | [jchelper 0.1.0](https://github.com/CCBR/journal-club/releases/tag/v0.1.0) | 2024-05-07 | 2 | +| [reports](https://github.com/CCBR/reports) | [ccbr.reports 0.2.0](https://github.com/CCBR/reports/releases/tag/v0.2.0) | 2024-04-30 | 11 | +| [parkit](https://github.com/CCBR/parkit) | [v2.0.1](https://github.com/CCBR/parkit/releases/tag/v2.0.1) | 2024-04-16 | 0 | +| [CCBR_tobias](https://github.com/CCBR/CCBR_tobias) | [CCBR_tobias 0.3.0](https://github.com/CCBR/CCBR_tobias/releases/tag/v0.3.0) | 2024-04-12 | 1 | +| [RENEE](https://github.com/CCBR/RENEE) | [RENEE 2.5.12](https://github.com/CCBR/RENEE/releases/tag/v2.5.12) | 2024-04-12 | 22 | +| [METRO](https://github.com/CCBR/METRO) | [v2.1](https://github.com/CCBR/METRO/releases/tag/v2.1) | 2024-03-28 | 2 | +| [CCBR-1144](https://github.com/CCBR/CCBR-1144) | [Data Release Latest](https://github.com/CCBR/CCBR-1144/releases/tag/v1.0.0) | 2024-03-04 | 0 | +| [CARLISLE](https://github.com/CCBR/CARLISLE) | [v2.5.0](https://github.com/CCBR/CARLISLE/releases/tag/v.2.5.0) | 2024-02-26 | 13 | +| [TRANQUIL](https://github.com/CCBR/TRANQUIL) | [TRANQUIL 0.2.1](https://github.com/CCBR/TRANQUIL/releases/tag/v0.2.1) | 2024-02-22 | 0 | +| [ccbr1271_ERVpipeline](https://github.com/CCBR/ccbr1271_ERVpipeline) | [v1.0.3](https://github.com/CCBR/ccbr1271_ERVpipeline/releases/tag/v1.0.3) | 2024-02-21 | 1 | +| [nf-sandbox](https://github.com/CCBR/nf-sandbox) | [nf-sandbox 0.2.1](https://github.com/CCBR/nf-sandbox/releases/tag/v0.2.1) | 2024-01-26 | 3 | +| [CHAMPAGNE](https://github.com/CCBR/CHAMPAGNE) | [CHAMPAGNE 0.3.0](https://github.com/CCBR/CHAMPAGNE/releases/tag/v0.3.0) | 2024-01-18 | 30 | +| [ASPEN](https://github.com/CCBR/ASPEN) | [v1.0.1](https://github.com/CCBR/ASPEN/releases/tag/v1.0.1) | 2023-12-27 | 6 | +| [CHARLIE](https://github.com/CCBR/CHARLIE) | [v0.10.1](https://github.com/CCBR/CHARLIE/releases/tag/v0.10.1) | 2023-12-23 | 20 | +| [nf-modules](https://github.com/CCBR/nf-modules) | [nf-modules 0.1.0](https://github.com/CCBR/nf-modules/releases/tag/v0.1.0) | 2023-11-29 | 11 | +| [CRISPIN](https://github.com/CCBR/CRISPIN) | [CRUISE 0.1.1](https://github.com/CCBR/CRISPIN/releases/tag/v0.1.1) | 2023-11-06 | 14 | +| [SINCLAIR](https://github.com/CCBR/SINCLAIR) | [v0.2.0](https://github.com/CCBR/SINCLAIR/releases/tag/v0.2.0) | 2023-11-01 | 26 | +| [CRISPRAnnotation](https://github.com/CCBR/CRISPRAnnotation) | [Code/Data Release](https://github.com/CCBR/CRISPRAnnotation/releases/tag/v1.0) | 2023-10-19 | 0 | +| [SharanLab](https://github.com/CCBR/SharanLab) | [Data/Code Release](https://github.com/CCBR/SharanLab/releases/tag/v1.0.0) | 2023-07-18 | 0 | +| [Pipeliner](https://github.com/CCBR/Pipeliner) | [v4.0.7](https://github.com/CCBR/Pipeliner/releases/tag/v4.0.7) | 2023-05-09 | 24 | +| [MAPLE](https://github.com/CCBR/MAPLE) | [version 1.0.1](https://github.com/CCBR/MAPLE/releases/tag/v1.0.1) | 2023-02-27 | 0 | +| [DTB_ExomeSeq](https://github.com/CCBR/DTB_ExomeSeq) | [v1.0](https://github.com/CCBR/DTB_ExomeSeq/releases/tag/v1.0) | 2022-06-22 | 0 | +| [ASCENT](https://github.com/CCBR/ASCENT) | [v0.1.1](https://github.com/CCBR/ASCENT/releases/tag/v0.1.1) | 2022-01-04 | 2 | +| [Antitumor-activity-of-entinostat-plus-NHS-IL12](https://github.com/CCBR/Antitumor-activity-of-entinostat-plus-NHS-IL12) | [Manuscript Methods](https://github.com/CCBR/Antitumor-activity-of-entinostat-plus-NHS-IL12/releases/tag/v.1.0) | 2021-06-01 | 0 | +| [CCBR_circRNA_AmpliconSeq](https://github.com/CCBR/CCBR_circRNA_AmpliconSeq) | [v0.1.1](https://github.com/CCBR/CCBR_circRNA_AmpliconSeq/releases/tag/v0.1.1) | 2021-03-24 | 0 | +| [rNA](https://github.com/CCBR/rNA) | [Release v1.0.0](https://github.com/CCBR/rNA/releases/tag/v1.0.0) | 2021-01-21 | 0 | +| [l2p](https://github.com/CCBR/l2p) | [Release v0.0.3](https://github.com/CCBR/l2p/releases/tag/v0.0.3) | 2020-07-13 | 0 | +| [MAAPster](https://github.com/CCBR/MAAPster) | [Release v2.0.0](https://github.com/CCBR/MAAPster/releases/tag/v2.0.0) | 2020-04-27 | 0 | +| [ChIP-Seq-Pipeline](https://github.com/CCBR/ChIP-Seq-Pipeline) | [Alpha 2](https://github.com/CCBR/ChIP-Seq-Pipeline/releases/tag/alpha2) | 2016-10-01 | 2 |+ Back to Top +